PENGUINN: Precise Exploration of Nuclear G-Quadruplexes Using Interpretable Neural Networks
- PMID: 33193663
- PMCID: PMC7653191
- DOI: 10.3389/fgene.2020.568546
PENGUINN: Precise Exploration of Nuclear G-Quadruplexes Using Interpretable Neural Networks
Abstract
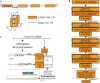
G-quadruplexes (G4s) are a class of stable structural nucleic acid secondary structures that are known to play a role in a wide spectrum of genomic functions, such as DNA replication and transcription. The classical understanding of G4 structure points to four variable length guanine strands joined by variable length nucleotide stretches. Experiments using G4 immunoprecipitation and sequencing experiments have produced a high number of highly probable G4 forming genomic sequences. The expense and technical difficulty of experimental techniques highlights the need for computational approaches of G4 identification. Here, we present PENGUINN, a machine learning method based on Convolutional neural networks, that learns the characteristics of G4 sequences and accurately predicts G4s outperforming state-of-the-art methods. We provide both a standalone implementation of the trained model, and a web application that can be used to evaluate sequences for their G4 potential.
Keywords: G quadruplex; bioinformatics and computational biology; deep neural network; genomic; imbalanced data classification; machine learning; web application.
Copyright © 2020 Klimentova, Polacek, Simecek and Alexiou.
Figures

References
-
- Barshai M., Orenstein Y. (2019). “Predicting G-quadruplexes from DNA sequences using multi-kernel convolutional neural networks,” in Proceedings of the 10th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics - BCB ’19 New York, NY.
LinkOut - more resources
Full Text Sources

