Integrated transcriptome meta-analysis of pancreatic ductal adenocarcinoma and matched adjacent pancreatic tissues
- PMID: 33194391
- PMCID: PMC7597628
- DOI: 10.7717/peerj.10141
Integrated transcriptome meta-analysis of pancreatic ductal adenocarcinoma and matched adjacent pancreatic tissues
Abstract
A comprehensive meta-analysis of publicly available gene expression microarray data obtained from human-derived pancreatic ductal adenocarcinoma (PDAC) tissues and their histologically matched adjacent tissue samples was performed to provide diagnostic and prognostic biomarkers, and molecular targets for PDAC. An integrative meta-analysis of four submissions (GSE62452, GSE15471, GSE62165, and GSE56560) containing 105 eligible tumor-adjacent tissue pairs revealed 344 differentially over-expressed and 168 repressed genes in PDAC compared to the adjacent-to-tumor samples. The validation analysis using TCGA combined GTEx data confirmed 98.24% of the identified up-regulated and 73.88% of the down-regulated protein-coding genes in PDAC. Pathway enrichment analysis showed that "ECM-receptor interaction", "PI3K-Akt signaling pathway", and "focal adhesion" are the most enriched KEGG pathways in PDAC. Protein-protein interaction analysis identified FN1, TIMP1, and MSLN as the most highly ranked hub genes among the DEGs. Transcription factor enrichment analysis revealed that TCF7, CTNNB1, SMAD3, and JUN are significantly activated in PDAC, while SMAD7 is inhibited. The prognostic significance of the identified and validated differentially expressed genes in PDAC was evaluated via survival analysis of TCGA Pan-Cancer pancreatic ductal adenocarcinoma data. The identified candidate prognostic biomarkers were then validated in four external validation datasets (GSE21501, GSE50827, GSE57495, and GSE71729) to further improve reliability. A total of 28 up-regulated genes were found to be significantly correlated with worse overall survival in patients with PDAC. Twenty-one of the identified prognostic genes (ITGB6, LAMC2, KRT7, SERPINB5, IGF2BP3, IL1RN, MPZL2, SFTA2, MET, LAMA3, ARNTL2, SLC2A1, LAMB3, COL17A1, EPSTI1, IL1RAP, AK4, ANXA2, S100A16, KRT19, and GPRC5A) were also found to be significantly correlated with the pathological stages of the disease. The results of this study provided promising prognostic biomarkers that have the potential to differentiate PDAC from both healthy and adjacent-to-tumor pancreatic tissues. Several novel dysregulated genes merit further study as potentially promising candidates for the development of more effective treatment strategies for PDAC.
Keywords: Biomarker; Gene expression; Gene expression omnibus; Microarray; Pancreatic ductal adenocarcinoma.
© 2020 Atay.
Conflict of interest statement
The author declares that they have no competing interests.
Figures
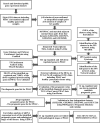
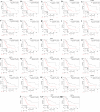
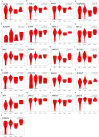

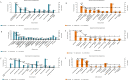
References
-
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. Nature Genetics. 2000;25:25–29. doi: 10.1038/75556. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

