Differentially Methylated Regions in Desmoid-Type Fibromatosis: A Comparison Between CTNNB1 S45F and T41A Tumors
- PMID: 33194643
- PMCID: PMC7658920
- DOI: 10.3389/fonc.2020.565031
Differentially Methylated Regions in Desmoid-Type Fibromatosis: A Comparison Between CTNNB1 S45F and T41A Tumors
Abstract
Introduction: The majority of desmoid-type fibromatosis (DTF) tumors harbor a β-catenin mutation, affecting specific codons in CTNNB1 exon 3. S45F tumors are reported to have a higher chance of recurrence after surgery and more resistance to systemic treatments compared to wild-type (WT) and T41A tumors. The aim of this pilot study was to examine the genome-wide DNA methylation profiles of S45F and T41A mutated DTF, to explain the observed differences in clinical behavior between these DTF subtypes.
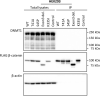
Material and methods: Genome-wide analysis of DNA methylation was performed using MeD-seq on formalin-fixed, paraffin-embedded primary DTF samples harboring a S45F (n = 14) or a T41A (n = 15) mutation. Differentially methylated regions (DMRs) between S45F and T41A DTF were identified and used for a supervised hierarchical cluster analysis. DMRs with a fold-change ≥1.5 were considered to be differentially methylated and differences between S45F and T41A tumors were quantitatively assessed. The effect of DMRs on the expression of associated genes was assessed using an independent mRNA expression dataset. Protein-protein interactions between WT β-catenin and mutant variants and DNA methyltransferase 1 (DNMT1) were examined by immunoprecipitation experiments.
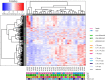
Results: MeD-seq analyses indicated 354 regions that displayed differential methylation. Cluster analysis yielded no distinct clusters based on mutation, sex, tumor site or tumor size. A supervised clustering based on DMRs between small (≤34 mm) and large (>87 mm) DTF distinguished the two groups. Only ten DMRs displayed a fold change of ≥1.5 and six of them were found associated with the following genes: NLRP4, FOXK2, PERM1, CCDC6, NOC4L, and DUX4L6. The effects of DMRs on gene expression yielded a significant difference (p < 0.05) in the expression between S45F and T41A for CCDC6 and FOXK2 but not for all Affymetrix probe-sets used to detect these genes. Immunoprecipitations did not reveal an association of WT β-catenin or mutant variants with DNMT1.
Conclusion: This study demonstrated that S45F and T41A DTF tumors did not exhibit gross differences in DNA methylation patterns. This implies that distinct DNA methylation profiles are not the sole determinant for the divergent clinical behavior of these different DTF mutant subtypes.
Keywords: DNA methylation; aggressive fibromatosis; epigenomics; soft tissue sarcomas; β-catenin.
Copyright © 2020 Timbergen, Boers, Vriends, Boers, van IJcken, Lavrijsen, Grünhagen, Verhoef, Sleijfer, Smits, Gribnau and Wiemer.
Figures

References
-
- Fletcher CDM, Bridge JA, Hogendoorn PCW, Mertens F. World Health Organization, International Agency for Research on Cancer. WHO classification of tumours of soft tissue and bone. Press: Lyon: IARC; (2013). p. 2013.
-
- Crago AM, Chmielecki J, Rosenberg M, O’Connor R, Byrne C, Wilder FG, et al. Near universal detection of alterations in CTNNB1 and Wnt pathway regulators in desmoid-type fibromatosis by whole-exome sequencing and genomic analysis. Genes Chromosomes Cancer (2015) 54(10):606–15. 10.1002/gcc.22272 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

