Challenges and Advances in Antemortem Diagnosis of Human Transmissible Spongiform Encephalopathies
- PMID: 33195151
- PMCID: PMC7606880
- DOI: 10.3389/fbioe.2020.585896
Challenges and Advances in Antemortem Diagnosis of Human Transmissible Spongiform Encephalopathies
Abstract
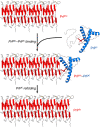
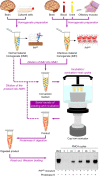
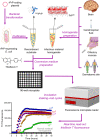
Transmissible spongiform encephalopathies (TSEs), also known as prion diseases, arise from the structural conversion of the monomeric, cellular prion protein (PrPC) into its multimeric scrapie form (PrPSc). These pathologies comprise a group of intractable, rapidly evolving neurodegenerative diseases. Currently, a definitive diagnosis of TSE relies on the detection of PrPSc and/or the identification of pathognomonic histological features in brain tissue samples, which are usually obtained postmortem or, in rare cases, by brain biopsy (antemortem). Over the past two decades, several paraclinical tests for antemortem diagnosis have been developed to preclude the need for brain samples. Some of these alternative methods have been validated and can provide a probable diagnosis when combined with clinical evaluation. Paraclinical tests include in vitro cell-free conversion techniques, such as the real-time quaking-induced conversion (RT-QuIC), as well as immunoassays, electroencephalography (EEG), and brain bioimaging methods, such as magnetic resonance imaging (MRI), whose importance has increased over the years. PrPSc is the main biomarker in TSEs, and the RT-QuIC assay stands out for its ability to detect PrPSc in cerebrospinal fluid (CSF), olfactory mucosa, and dermatome skin samples with high sensitivity and specificity. Other biochemical biomarkers are the proteins 14-3-3, tau, neuron-specific enolase (NSE), astroglial protein S100B, α-synuclein, and neurofilament light chain protein (NFL), but they are not specific for TSEs. This paper reviews the techniques employed for definite diagnosis, as well as the clinical and paraclinical methods for possible and probable diagnosis, both those in use currently and those no longer employed. We also discuss current criteria, challenges, and perspectives for TSE diagnosis. An early and accurate diagnosis may allow earlier implementation of strategies to delay or stop disease progression.
Keywords: cell-free conversion assay; diagnostic; neurodegenerative disease; prion diseases; prion protein.
Copyright © 2020 Ascari, Rocha, Gonçalves, Vieira and Cordeiro.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

