Evaluation of CONSRANK-Like Scoring Functions for Rescoring Ensembles of Protein-Protein Docking Poses
- PMID: 33195406
- PMCID: PMC7641601
- DOI: 10.3389/fmolb.2020.559005
Evaluation of CONSRANK-Like Scoring Functions for Rescoring Ensembles of Protein-Protein Docking Poses
Abstract
Scoring is a challenging step in protein-protein docking, where typically thousands of solutions are generated. In this study, we ought to investigate the contribution of consensus-rescoring, as introduced by Oliva et al. (2013) with the CONSRANK method, where the set of solutions is used to build statistics in order to identify recurrent solutions. We explore several ways to perform consensus-based rescoring on the ZDOCK decoy set for Benchmark 4. We show that the information of the interface size is critical for successful rescoring in this context, but that consensus rescoring in itself performs less well than traditional physics-based evaluation. The results of physics-based and consensus-based rescoring are partially overlapping, supporting the use of a combination of these approaches.
Keywords: docking; interface; prediction; protein–protein interaction; scoring.
Copyright © 2020 Launay, Ohue, Prieto Santero, Matsuzaki, Hilpert, Uchikoga, Hayashi and Martin.
Figures

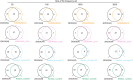
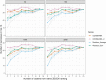

References
LinkOut - more resources
Full Text Sources
Other Literature Sources

