N5 Is the New C4a: Biochemical Functionalization of Reduced Flavins at the N5 Position
- PMID: 33195440
- PMCID: PMC7662398
- DOI: 10.3389/fmolb.2020.598912
N5 Is the New C4a: Biochemical Functionalization of Reduced Flavins at the N5 Position
Abstract
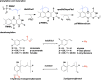
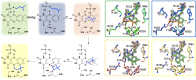
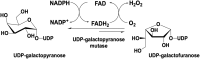
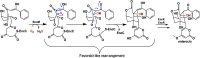
For three decades the C4a-position of reduced flavins was the known site for covalency within flavoenzymes. The reactivity of this position of the reduced isoalloxazine ring with the dioxygen ground-state triplet established the C4a as a site capable of one-electron chemistry. Within the last two decades new types of reduced flavin reactivity have been documented. These studies reveal that the N5 position is also a protean site of reactivity, that is capable of nucleophilic attack to form covalent bonds with substrates. In addition, though the precise mechanism of dioxygen reactivity is yet to be definitively demonstrated, it is clear that the N5 position is directly involved in substrate oxygenation in some enzymes. In this review we document the lineage of discoveries that identified five unique modes of N5 reactivity that collectively illustrate the versatility of this position of the reduced isoalloxazine ring.
Keywords: C4a-position; N5-position; covalent; flavin; imino; oxide; peroxo.
Copyright © 2020 Beaupre and Moran.
Figures









Similar articles
-
QM/MM Modeling of the Flavin Functionalization in the RutA Monooxygenase.Molecules. 2023 Mar 6;28(5):2405. doi: 10.3390/molecules28052405. Molecules. 2023. PMID: 36903648 Free PMC article.
-
Proton-coupled electron transfer and adduct configuration are important for C4a-hydroperoxyflavin formation and stabilization in a flavoenzyme.J Am Chem Soc. 2014 Jan 8;136(1):241-53. doi: 10.1021/ja4088055. Epub 2013 Dec 24. J Am Chem Soc. 2014. PMID: 24368083
-
Mechanism of Oxygen Activation in a Flavin-Dependent Monooxygenase: A Nearly Barrierless Formation of C4a-Hydroperoxyflavin via Proton-Coupled Electron Transfer.J Am Chem Soc. 2015 Jul 29;137(29):9363-74. doi: 10.1021/jacs.5b04328. Epub 2015 Jul 20. J Am Chem Soc. 2015. PMID: 26144862
-
Sweating the assets of flavin cofactors: new insight of chemical versatility from knowledge of structure and mechanism.Curr Opin Struct Biol. 2016 Dec;41:19-26. doi: 10.1016/j.sbi.2016.05.014. Epub 2016 Jun 3. Curr Opin Struct Biol. 2016. PMID: 27266331 Review.
-
Insights into the enzymatic formation, chemical features, and biological role of the flavin-N5-oxide.Curr Opin Chem Biol. 2018 Dec;47:47-53. doi: 10.1016/j.cbpa.2018.08.003. Epub 2018 Aug 27. Curr Opin Chem Biol. 2018. PMID: 30165331 Review.
Cited by
-
DszA Catalyzes C-S Bond Cleavage through N5-Hydroperoxyl Formation.J Chem Inf Model. 2024 May 27;64(10):4218-4230. doi: 10.1021/acs.jcim.4c00301. Epub 2024 Apr 29. J Chem Inf Model. 2024. PMID: 38684937 Free PMC article.
-
Structural insights into the initiation of free radical formation in the Class Ib ribonucleotide reductases in Mycobacteria.Curr Res Struct Biol. 2024 Sep 18;8:100157. doi: 10.1016/j.crstbi.2024.100157. eCollection 2024. Curr Res Struct Biol. 2024. PMID: 39399574 Free PMC article.
-
Determining large hyperfine interactions of a model flavoprotein in the semiquinone state using pulse EPR (electron paramagnetic resonance) techniques.Magn Reson (Gott). 2025 Jul 17;6(2):183-197. doi: 10.5194/mr-6-183-2025. eCollection 2025. Magn Reson (Gott). 2025. PMID: 40771403 Free PMC article.
-
Biosynthetic Strategies of Berberine Bridge Enzyme-like Flavoprotein Oxidases toward Structural Diversification in Natural Product Biosynthesis.Biochemistry. 2024 Sep 3;63(17):2089-2110. doi: 10.1021/acs.biochem.4c00320. Epub 2024 Aug 12. Biochemistry. 2024. PMID: 39133819 Free PMC article. Review.
-
Structural Basis of Sequential Enantioselective Epoxidation by a Flavin-Dependent Monooxygenase in Lasalocid A Biosynthesis.Angew Chem Int Ed Engl. 2025 Jun 10;64(24):e202504982. doi: 10.1002/anie.202504982. Epub 2025 Apr 26. Angew Chem Int Ed Engl. 2025. PMID: 40199722 Free PMC article.
References
-
- Alston T. A., Porter D. J., Bright H. J. (1983). Suicide inactivation of D-amino acid oxidase by 1-chloro-1-nitroethane. J. Biol. Chem. 258 1136–1141. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

