centrocortin RNA localization to centrosomes is regulated by FMRP and facilitates error-free mitosis
- PMID: 33196763
- PMCID: PMC7716377
- DOI: 10.1083/jcb.202004101
centrocortin RNA localization to centrosomes is regulated by FMRP and facilitates error-free mitosis
Abstract
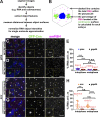
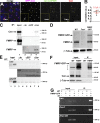
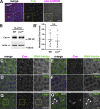
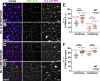
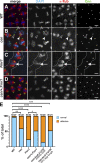
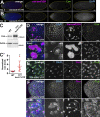
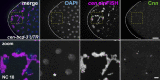
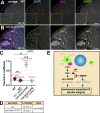
Centrosomes are microtubule-organizing centers required for error-free mitosis and embryonic development. The microtubule-nucleating activity of centrosomes is conferred by the pericentriolar material (PCM), a composite of numerous proteins subject to cell cycle-dependent oscillations in levels and organization. In diverse cell types, mRNAs localize to centrosomes and may contribute to changes in PCM abundance. Here, we investigate the regulation of mRNA localization to centrosomes in the rapidly cycling Drosophila melanogaster embryo. We find that RNA localization to centrosomes is regulated during the cell cycle and developmentally. We identify a novel role for the fragile-X mental retardation protein in the posttranscriptional regulation of a model centrosomal mRNA, centrocortin (cen). Further, mistargeting cen mRNA is sufficient to alter cognate protein localization to centrosomes and impair spindle morphogenesis and genome stability.
© 2020 Ryder et al.
Figures




Comment in
-
Think globally, act locally: Centrosome-localized mRNAs ensure mitotic fidelity.J Cell Biol. 2020 Dec 7;219(12):e202010172. doi: 10.1083/jcb.202010172. J Cell Biol. 2020. PMID: 33216116 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

