Epigenome engineering: new technologies for precision medicine
- PMID: 33196851
- PMCID: PMC7736826
- DOI: 10.1093/nar/gkaa1000
Epigenome engineering: new technologies for precision medicine
Abstract
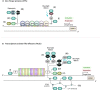
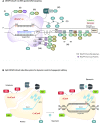
Chromatin adopts different configurations that are regulated by reversible covalent modifications, referred to as epigenetic marks. Epigenetic inhibitors have been approved for clinical use to restore epigenetic aberrations that result in silencing of tumor-suppressor genes, oncogene addictions, and enhancement of immune responses. However, these drugs suffer from major limitations, such as a lack of locus selectivity and potential toxicities. Technological advances have opened a new era of precision molecular medicine to reprogram cellular physiology. The locus-specificity of CRISPR/dCas9/12a to manipulate the epigenome is rapidly becoming a highly promising strategy for personalized medicine. This review focuses on new state-of-the-art epigenome editing approaches to modify the epigenome of neoplasms and other disease models towards a more 'normal-like state', having characteristics of normal tissue counterparts. We highlight biomolecular engineering methodologies to assemble, regulate, and deliver multiple epigenetic effectors that maximize the longevity of the therapeutic effect, and we discuss limitations of the platforms such as targeting efficiency and intracellular delivery for future clinical applications.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

