Stability of Human Serum Amyloid A Fibrils
- PMID: 33197318
- PMCID: PMC7688565
- DOI: 10.1021/acs.jpcb.0c08280
Stability of Human Serum Amyloid A Fibrils
Abstract
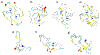
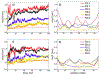
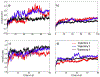
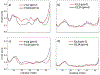
In systemic amyloidosis, serum amyloid A (SAA) fibril deposits cause widespread damages to tissues and organs that eventually may lead to death. A therapeutically intervention therefore has either to dissolve these fibrils or inhibit their formation. However, only recently has the human SAA fibril structure been resolved at a resolution that is sufficient for development of drug candidates. Here, we use molecular dynamic simulations to probe the factors that modulate the stability of this fibril model. Our simulations suggest that fibril formation starts with the stacking of two misfolded monomers into metastable dimers, with the stacking depending on the N-terminal amyloidogenic regions of different chains forming anchors. The resulting dimers pack in a second step into a 2-fold two-layer tetramer that is stable enough to nucleate fibril formation. The stability of the initial dimers is enhanced under acidic conditions by a strong salt bridge and side-chain hydrogen bond network in the C-terminal cavity (residues 23-51) but is not affected by the presence of the disordered C-terminal tail.
Figures
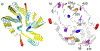

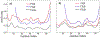
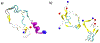
References
-
- Qi R; Luo Y; Wei G; Nussinov R; Ma B, Aβ “stretching-and-packing” cross-seeding mechanism can trigger tau protein aggregation. The Journal of Physical Chemistry Letters 2015, 6 (16), 3276–3282.
-
- Sipe JD; Benson MD; Buxbaum JN; Ikeda S.-i.; Merlini G; Saraiva MJ; Westermark P, Amyloid fibril proteins and amyloidosis: chemical identification and clinical classification International Society of Amyloidosis 2016 Nomenclature Guidelines. Amyloid 2016, 23 (4), 209–213. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

