E3 Ubiquitin Ligase TRIP12: Regulation, Structure, and Physiopathological Functions
- PMID: 33198194
- PMCID: PMC7697007
- DOI: 10.3390/ijms21228515
E3 Ubiquitin Ligase TRIP12: Regulation, Structure, and Physiopathological Functions
Abstract
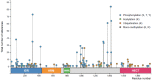
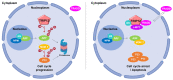
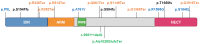
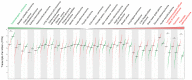
The Thyroid hormone Receptor Interacting Protein 12 (TRIP12) protein belongs to the 28-member Homologous to the E6-AP C-Terminus (HECT) E3 ubiquitin ligase family. First described as an interactor of the thyroid hormone receptor, TRIP12's biological importance was revealed by the embryonic lethality of a murine model bearing an inactivating mutation in the TRIP12 gene. Further studies showed the participation of TRIP12 in the regulation of major biological processes such as cell cycle progression, DNA damage repair, chromatin remodeling, and cell differentiation by an ubiquitination-mediated degradation of key protein substrates. Moreover, alterations of TRIP12 expression have been reported in cancers that can serve as predictive markers of therapeutic response. The TRIP12 gene is also referenced as a causative gene associated to intellectual disorders such as Clark-Baraitser syndrome and is clearly implicated in Autism Spectrum Disorder. The aim of the review is to provide an exhaustive and integrated overview of the different aspects of TRIP12 ranging from its regulation, molecular functions and physio-pathological implications.
Keywords: E3 ubiquitin ligase; TRIP12; cancers; intellectual disorders.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
-
- Nomura N., Nagase T., Miyajima N., Sazuka T., Tanaka A., Sato S., Seki N., Kawarabayasi Y., Ishikawa K.-I., Tabata S. Prediction of the Coding Sequences of Unidentified Human Genes. II. The Coding Sequences of 40 New Genes (KIAA0041-KIAA0080) Deduced by Analysis of cDNA Clones from Human Cell Line KG-1. DNA Res. 1994;1:223–229. doi: 10.1093/dnares/1.5.223. - DOI - PubMed
-
- Lee J.W., Choi H.S., Gyuris J., Brent R., Moore D.D. Two classes of proteins dependent on either the presence or absence of thyroid hormone for interaction with the thyroid hormone receptor. Mol. Endocrinol. 1995;9:243–254. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

