Soybean Viromes in the Republic of Korea Revealed by RT-PCR and Next-Generation Sequencing
- PMID: 33198273
- PMCID: PMC7698195
- DOI: 10.3390/microorganisms8111777
Soybean Viromes in the Republic of Korea Revealed by RT-PCR and Next-Generation Sequencing
Abstract
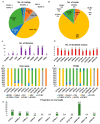
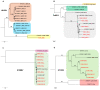
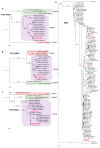
Soybean (Glycine max L.) is one of the most important crop plants in the Republic of Korea. Here, we conducted a soybean virome study. We harvested a total of 172 soybean leaf samples showing disease symptoms from major soybean-growing regions in the Republic of Korea. Individual samples were examined for virus infection by RT-PCR. Moreover, we generated eight libraries representing eight provinces by pooling samples and four libraries from single samples. RNA-seq followed by bioinformatics analyses revealed 10 different RNA viruses infecting soybean. The proportion of viral reads in each transcriptome ranged from 0.2 to 31.7%. Coinfection of different viruses in soybean plants was very common. There was a single dominant virus in each province, and this geographical difference might be related to the soybean seeds that transmit viruses. In this study, 32 viral genome sequences were assembled and successfully used to analyze the phylogenetic relationships and quasispecies nature of the identified RNA viruses. Moreover, RT-PCR with newly developed primers confirmed infection of the identified viruses in each library. Taken together, our soybean virome study provides a comprehensive overview of viruses infecting soybean in eight geographical regions in the Republic of Korea and four single soybean plants in detail.
Keywords: Korea; RNA-seq; mutation; soybean; viral genome; virome; virus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Hill J.H., Whitham S.A. Advances in Virus Research. Volume 90. Elsevier; Amsterdam, The Netherlands: 2014. Control of virus diseases in soybeans; pp. 355–390. - PubMed
-
- Cho E.-K., Goodman R.M. Strains of soybean mosaic virus: Classification based on virulence in resistant soybean cultivars. Phytopathology. 1979;69:467–470. doi: 10.1094/Phyto-69-467. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

