History of the COVID-19 pandemic: Origin, explosion, worldwide spreading
- PMID: 33199023
- PMCID: PMC7834510
- DOI: 10.1016/j.bbrc.2020.10.087
History of the COVID-19 pandemic: Origin, explosion, worldwide spreading
Abstract
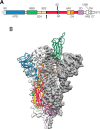
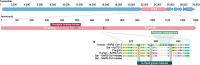
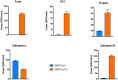
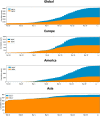
The SARS-CoV-2 virus of the COVID-19 pandemic, that is presently devastating the entire world, had been active well before January of this year, when its pathogenic potential exploded full force in Wuhan. It had caused the onset of small disease outbreaks in China, and probably elsewhere as well, which failed to reach epidemic potential. The distant general origin of its zoonosis can be traced back to the ecosystem changes that have decreased biodiversity, greatly facilitating the contacts between humans and the animal reservoirs that carry pathogens, including SARS-CoV-2. These reservoirs are the bats. The transition between the limited outbreaks that had occurred through 2019 and the epidemic explosion of December-January was made possible by the great amplification of the general negative conditions that had caused the preceding small outbreaks. In the light of what we have now learned, the explosion was predictable, and could have happened wherever the conditions that had allowed it, could be duplicated. What could not have been predicted was the second transition, from epidemic to pandemic. Research has now revealed that the globalization of the infection appears to have been caused by a mutation in the spike protein of the SARS-CoV-2, that has dramatically increased its transmissibility.
Keywords: Adenosine analogs; Antiviral therapy; RNA dependent RNA polymerase; Remdesivir.
Copyright © 2020. Published by Elsevier Inc.
Conflict of interest statement
Declaration of competing interest The authors declare that they don’t have conficts of interest.
Figures
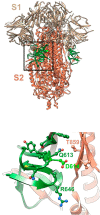
References
-
- Luis A.D., Hayman D.T.S., O’Shea T.J., Cryan P.M., Gilbert A.T., Pulliam J.R.C., Mills J.N., Timonin M.E., Willis C.K.R., Cunningham A.A., Fooks A.R., Rupprecht C.E., Wood J.L.N., Webb C.T. A comparison of bats and rodents as reservoirs of zoonotic viruses: are bats special ? Proc. Royal Soc. B. 2013;280 doi: 10.1098/rspb.2012.2753. Print 2013 Apr 7. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

