The KH domain facilitates the substrate specificity and unwinding processivity of DDX43 helicase
- PMID: 33199368
- PMCID: PMC7949032
- DOI: 10.1074/jbc.RA120.015824
The KH domain facilitates the substrate specificity and unwinding processivity of DDX43 helicase
Abstract
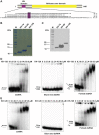
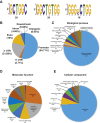
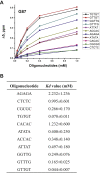
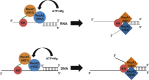
The K-homology (KH) domain is a nucleic acid-binding domain present in many proteins. Recently, we found that the DEAD-box helicase DDX43 contains a KH domain in its N-terminus; however, its function remains unknown. Here, we purified recombinant DDX43 KH domain protein and found that it prefers binding ssDNA and ssRNA. Electrophoretic mobility shift assay and NMR revealed that the KH domain favors pyrimidines over purines. Mutational analysis showed that the GXXG loop in the KH domain is involved in pyrimidine binding. Moreover, we found that an alanine residue adjacent to the GXXG loop is critical for binding. Systematic evolution of ligands by exponential enrichment, chromatin immunoprecipitation-seq, and cross-linking immunoprecipitation-seq showed that the KH domain binds C-/T-rich DNA and U-rich RNA. Bioinformatics analysis suggested that the KH domain prefers to bind promoters. Using 15N-heteronuclear single quantum coherence NMR, the optimal binding sequence was identified as TTGT. Finally, we found that the full-length DDX43 helicase prefers DNA or RNA substrates with TTGT or UUGU single-stranded tails and that the KH domain is critically important for sequence specificity and unwinding processivity. Collectively, our results demonstrated that the KH domain facilitates the substrate specificity and processivity of the DDX43 helicase.
Keywords: CLIP-seq; ChIP-seq; DDX43; KH domain; NMR; SELEX; helicase processivity; substrate specificity.
Copyright © 2020 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures





References
-
- Linder P., Jankowsky E. From unwinding to clamping - the DEAD box RNA helicase family. Nat. Rev. Mol. Cell Biol. 2011;12:505–516. - PubMed
-
- Lohman T.M., Bjornson K.P. Mechanisms of helicase-catalyzed DNA unwinding. Annu. Rev. Biochem. 1996;65:169–214. - PubMed
-
- Pyle A.M. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu. Rev. Biophys. 2008;37:317–336. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

