Glutaric acid production by systems metabolic engineering of an l-lysine-overproducing Corynebacterium glutamicum
- PMID: 33199604
- PMCID: PMC7720191
- DOI: 10.1073/pnas.2017483117
Glutaric acid production by systems metabolic engineering of an l-lysine-overproducing Corynebacterium glutamicum
Abstract
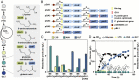
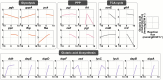
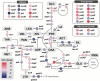
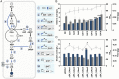
There is increasing industrial demand for five-carbon platform chemicals, particularly glutaric acid, a widely used building block chemical for the synthesis of polyesters and polyamides. Here we report the development of an efficient glutaric acid microbial producer by systems metabolic engineering of an l-lysine-overproducing Corynebacterium glutamicum BE strain. Based on our previous study, an optimal synthetic metabolic pathway comprising Pseudomonas putida l-lysine monooxygenase (davB) and 5-aminovaleramide amidohydrolase (davA) genes and C. glutamicum 4-aminobutyrate aminotransferase (gabT) and succinate-semialdehyde dehydrogenase (gabD) genes, was introduced into the C. glutamicum BE strain. Through system-wide analyses including genome-scale metabolic simulation, comparative transcriptome analysis, and flux response analysis, 11 target genes to be manipulated were identified and expressed at desired levels to increase the supply of direct precursor l-lysine and reduce precursor loss. A glutaric acid exporter encoded by ynfM was discovered and overexpressed to further enhance glutaric acid production. Fermentation conditions, including oxygen transfer rate, batch-phase glucose level, and nutrient feeding strategy, were optimized for the efficient production of glutaric acid. Fed-batch culture of the final engineered strain produced 105.3 g/L of glutaric acid in 69 h without any byproduct. The strategies of metabolic engineering and fermentation optimization described here will be useful for developing engineered microorganisms for the high-level bio-based production of other chemicals of interest to industry.
Keywords: Corynebacterium glutamicum; glutaric acid; metabolic engineering; multiomics.
Conflict of interest statement
The authors declare no competing interest.
Figures
References
-
- Choi K. R., et al. , Systems metabolic engineering strategies: Integrating systems and synthetic biology with metabolic engineering. Trends Biotechnol. 37, 817–837 (2019). - PubMed
-
- Lee S. Y., Kim H. U., Systems strategies for developing industrial microbial strains. Nat. Biotechnol. 33, 1061–1072 (2015). - PubMed
-
- Nielsen J., Keasling J. D., Engineering cellular metabolism. Cell 164, 1185–1197 (2016). - PubMed
-
- Atsumi S., Hanai T., Liao J. C., Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451, 86–89 (2008). - PubMed
-
- Lee S. Y., et al. , A comprehensive metabolic map for production of bio-based chemicals. Nat. Catal. 2, 18–33 (2019).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

