Incompatibility between proliferation and plant invasion is mediated by a regulator of appressorium formation in the corn smut fungus Ustilago maydis
- PMID: 33199618
- PMCID: PMC7720189
- DOI: 10.1073/pnas.2006909117
Incompatibility between proliferation and plant invasion is mediated by a regulator of appressorium formation in the corn smut fungus Ustilago maydis
Abstract
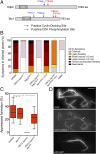
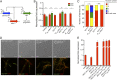
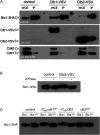
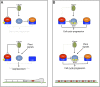
Plant pathogenic fungi often developed specialized infection structures to breach the outer surface of a host plant. These structures, called appressoria, lead the invasion of the plant by the fungal hyphae. Studies in different phytopathogenic fungi showed that appressorium formation seems to be subordinated to the cell cycle. This subordination ensures the loading in the invading hypha of the correct genetic information to proceed with plant infection. However, how the cell cycle transmits its condition to the genetic program controlling appressorium formation and promoting the plant's invasion is unknown. Our results have uncovered how this process occurs for the appressorium of Ustilago maydis, the agent responsible for corn smut disease. Here, we described that the complex Clb2-cyclin-dependent kinase (Cdk)1, one of the master regulators of G2/M cell cycle progression in U. maydis, interacts and controls the subcellular localization of Biz1, a transcriptional factor required for the activation of the appressorium formation. Besides, Biz1 can arrest the cell cycle by down-regulation of the gene encoding a second b-cyclin Clb1 also required for the G2/M transition. These results revealed a negative feedback loop between appressorium formation and cell cycle progression in U. maydis, which serves as a "toggle switch" to control the fungal decision between infecting the plant or proliferating out of the plant.
Keywords: appressorium; cell cycle regulation; corn smut; phytopathogenic fungi.
Conflict of interest statement
The authors declare no competing interest.
Figures


Similar articles
-
Appressorium formation in the corn smut fungus Ustilago maydis requires a G2 cell cycle arrest.Plant Signal Behav. 2015;10(4):e1001227. doi: 10.1080/15592324.2014.1001227. Plant Signal Behav. 2015. PMID: 25876077 Free PMC article.
-
Biz1, a zinc finger protein required for plant invasion by Ustilago maydis, regulates the levels of a mitotic cyclin.Plant Cell. 2006 Sep;18(9):2369-87. doi: 10.1105/tpc.106.042754. Epub 2006 Aug 11. Plant Cell. 2006. PMID: 16905655 Free PMC article.
-
Programmed cell cycle arrest is required for infection of corn plants by the fungus Ustilago maydis.Development. 2014 Dec;141(24):4817-26. doi: 10.1242/dev.113415. Epub 2014 Nov 19. Development. 2014. PMID: 25411209
-
[Parasitic strategy and regulation mechanism of Ustilago maydis - A review].Wei Sheng Wu Xue Bao. 2016 Sep;56(9):1385-97. Wei Sheng Wu Xue Bao. 2016. PMID: 29738207 Review. Chinese.
-
Cell biology of corn smut disease-Ustilago maydis as a model for biotrophic interactions.Curr Opin Microbiol. 2016 Dec;34:60-66. doi: 10.1016/j.mib.2016.07.020. Epub 2016 Aug 6. Curr Opin Microbiol. 2016. PMID: 27504540 Review.
Cited by
-
Appressoria-Small but Incredibly Powerful Structures in Plant-Pathogen Interactions.Int J Mol Sci. 2023 Jan 21;24(3):2141. doi: 10.3390/ijms24032141. Int J Mol Sci. 2023. PMID: 36768468 Free PMC article. Review.
-
SsCak1 Regulates Growth and Pathogenicity in Sclerotinia sclerotiorum.Int J Mol Sci. 2023 Aug 9;24(16):12610. doi: 10.3390/ijms241612610. Int J Mol Sci. 2023. PMID: 37628791 Free PMC article.
-
Comparative transcriptome profiling and weighted gene co-expression network analysis to identify core genes in maize (Zea mays L.) silks infected by multiple fungi.Front Plant Sci. 2022 Oct 27;13:985396. doi: 10.3389/fpls.2022.985396. eCollection 2022. Front Plant Sci. 2022. PMID: 36388593 Free PMC article.
References
-
- Schulze-Lefert P., Knocking on the heaven’s wall: Pathogenesis of and resistance to biotrophic fungi at the cell wall. Curr. Opin. Plant Biol. 7, 377–383 (2004). - PubMed
-
- Deising H. B., Werner S., Wernitz M., The role of fungal appressoria in plant infection. Microbes Infect./Inst. Pasteur 2, 1631–1641 (2000). - PubMed
-
- Mendgen K., Deising H., Infection structures of fungal plant pathogens -a cytological and physiological evaluation. New Phytol. 124, 193–213 (1993). - PubMed
-
- Mendgen K., Hahn M., Deising H., Morphogenesis and mechanisms of penetration by plant pathogenic fungi. Annu. Rev. Phytopathol. 34, 367–386 (1996). - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Miscellaneous

