Germline AGO2 mutations impair RNA interference and human neurological development
- PMID: 33199684
- PMCID: PMC7670403
- DOI: 10.1038/s41467-020-19572-5
Germline AGO2 mutations impair RNA interference and human neurological development
Abstract
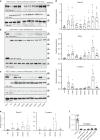
ARGONAUTE-2 and associated miRNAs form the RNA-induced silencing complex (RISC), which targets mRNAs for translational silencing and degradation as part of the RNA interference pathway. Despite the essential nature of this process for cellular function, there is little information on the role of RISC components in human development and organ function. We identify 13 heterozygous mutations in AGO2 in 21 patients affected by disturbances in neurological development. Each of the identified single amino acid mutations result in impaired shRNA-mediated silencing. We observe either impaired RISC formation or increased binding of AGO2 to mRNA targets as mutation specific functional consequences. The latter is supported by decreased phosphorylation of a C-terminal serine cluster involved in mRNA target release, increased formation of dendritic P-bodies in neurons and global transcriptome alterations in patient-derived primary fibroblasts. Our data emphasize the importance of gene expression regulation through the dynamic AGO2-RNA association for human neuronal development.
Conflict of interest statement
A.T. and K.M. are employees of GeneDx, Inc. The other authors declare that they have no conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

