Awakening a latent carbon fixation cycle in Escherichia coli
- PMID: 33199707
- PMCID: PMC7669889
- DOI: 10.1038/s41467-020-19564-5
Awakening a latent carbon fixation cycle in Escherichia coli
Abstract
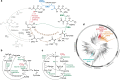
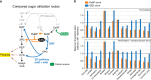
Carbon fixation is one of the most important biochemical processes. Most natural carbon fixation pathways are thought to have emerged from enzymes that originally performed other metabolic tasks. Can we recreate the emergence of a carbon fixation pathway in a heterotrophic host by recruiting only endogenous enzymes? In this study, we address this question by systematically analyzing possible carbon fixation pathways composed only of Escherichia coli native enzymes. We identify the GED (Gnd-Entner-Doudoroff) cycle as the simplest pathway that can operate with high thermodynamic driving force. This autocatalytic route is based on reductive carboxylation of ribulose 5-phosphate (Ru5P) by 6-phosphogluconate dehydrogenase (Gnd), followed by reactions of the Entner-Doudoroff pathway, gluconeogenesis, and the pentose phosphate pathway. We demonstrate the in vivo feasibility of this new-to-nature pathway by constructing E. coli gene deletion strains whose growth on pentose sugars depends on the GED shunt, a linear variant of the GED cycle which does not require the regeneration of Ru5P. Several metabolic adaptations, most importantly the increased production of NADPH, assist in establishing sufficiently high flux to sustain this growth. Our study exemplifies a trajectory for the emergence of carbon fixation in a heterotrophic organism and demonstrates a synthetic pathway of biotechnological interest.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

