A long-term study of AAV gene therapy in dogs with hemophilia A identifies clonal expansions of transduced liver cells
- PMID: 33199875
- PMCID: PMC7855056
- DOI: 10.1038/s41587-020-0741-7
A long-term study of AAV gene therapy in dogs with hemophilia A identifies clonal expansions of transduced liver cells
Abstract
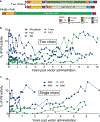
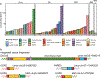
Nine dogs with hemophilia A were treated with adeno-associated viral (AAV) gene therapy and followed for up to 10 years. Administration of AAV8 or AAV9 vectors expressing canine factor VIII (AAV-cFVIII) corrected the FVIII deficiency to 1.9-11.3% of normal FVIII levels. In two of nine dogs, levels of FVIII activity increased gradually starting about 4 years after treatment. None of the dogs showed evidence of tumors or altered liver function. Analysis of integration sites in liver samples from six treated dogs identified 1,741 unique AAV integration events in genomic DNA and expanded cell clones in five dogs, with 44% of the integrations near genes involved in cell growth. All recovered integrated vectors were partially deleted and/or rearranged. Our data suggest that the increase in FVIII protein expression in two dogs may have been due to clonal expansion of cells harboring integrated vectors. These results support the clinical development of liver-directed AAV gene therapy for hemophilia A, while emphasizing the importance of long-term monitoring for potential genotoxicity.
Conflict of interest statement
Competing Interests Statement
D.E.S. receives royalites from a licensing agreement with Spark Therapeutics. D.E.S. and G.N.N. are inventors on a patent on factor VIII and hemophilia A gene therapy.
Figures




Comment in
-
Safety questions for AAV gene therapy.Nat Biotechnol. 2021 Jan;39(1):24-26. doi: 10.1038/s41587-020-00756-9. Nat Biotechnol. 2021. PMID: 33199877 No abstract available.
References
-
- Rangarajan S et al. AAV5-Factor VIII Gene Transfer in Severe Hemophilia A. N Engl J Med 377, 2519–2530 (2017). - PubMed
-
- High KA et al. A Phase 1/2 Trial of Investigational Spk-8011 in Hemophilia A Demonstrates Durable Expression and Prevention of Bleeds. in (ed. Blood) 132, 487 (2018).
-
- Nathwani AC et al. GO-8: Preliminary Results of a Phase I/II Dose Escalation Trial of Gene Therapy for Haemophilia A Using a Novel Human Factor VIII Variant. in (ed. Blood) 132, 489 (2018).
-
- Pasi KJ et al. Multiyear Follow-up of AAV5-hFVIII-SQ Gene Therapy for Hemophilia A. N Engl J Med 382, 29–40 (2020). - PubMed
-
- Jiang H et al. Multiyear therapeutic benefit of AAV serotypes 2, 6, and 8 delivering factor VIII to hemophilia A mice and dogs. Blood 108, 107–115 (2006). - PubMed
Methods-only References
-
- Hothorn T, Hornik K, van de Wiel M & Zeileis A Implementing a Class of Permutation Tests: The coin package. Journal of Statistical Software 28, 1–23 (2008).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

