Cell type-selective secretome profiling in vivo
- PMID: 33199915
- PMCID: PMC7904581
- DOI: 10.1038/s41589-020-00698-y
Cell type-selective secretome profiling in vivo
Abstract
Secreted polypeptides are a fundamental axis of intercellular and endocrine communication. However, a global understanding of the composition and dynamics of cellular secretomes in intact mammalian organisms has been lacking. Here, we introduce a proximity biotinylation strategy that enables labeling, detection and enrichment of secreted polypeptides in a cell type-selective manner in mice. We generate a proteomic atlas of hepatocyte, myocyte, pericyte and myeloid cell secretomes by direct purification of biotinylated secreted proteins from blood plasma. Our secretome dataset validates known cell type-protein pairs, reveals secreted polypeptides that distinguish between cell types and identifies new cellular sources for classical plasma proteins. Lastly, we uncover a dynamic and previously undescribed nutrient-dependent reprogramming of the hepatocyte secretome characterized by the increased unconventional secretion of the cytosolic enzyme betaine-homocysteine S-methyltransferase (BHMT). This secretome profiling strategy enables dynamic and cell type-specific dissection of the plasma proteome and the secreted polypeptides that mediate intercellular signaling.
Figures



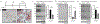

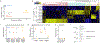
References
-
- Rorsman P & Braun M Regulation of insulin secretion in human pancreatic islets. Annu. Rev. Physiol 75, 155–179 (2013). - PubMed
-
- Gaceb A, Barbariga M, Özen I & Paul G The pericyte secretome: Potential impact on regeneration. Biochimie 155, 16–25 (2018). - PubMed
-
- Rabouille C Pathways of Unconventional Protein Secretion. Trends Cell Biol. 27, 230–240 (2017). - PubMed
-
- Eichelbaum K, Winter M, Diaz MB, Herzig S & Krijgsveld J Selective enrichment of newly synthesized proteins for quantitative secretome analysis. Nat. Biotechnol 30, 984–90 (2012). - PubMed
METHODS-ONLY REFERENCES
-
- Hailemariam M et al. S-Trap, an Ultrafast Sample-Preparation Approach for Shotgun Proteomics. J. Proteome Res. 17, 2917–2924 (2018). - PubMed
-
- Tyanova S, Temu T & Cox J The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc 11, 2301–2319 (2016). - PubMed
-
- Cox J et al. Andromeda: A peptide search engine integrated into the MaxQuant environment. J. Proteome Res 10, 1794–805 (2011). - PubMed
-
- Tyanova S et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–40 (2016). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

