In silico and in vitro studies reveal complement system drives coagulation cascade in SARS-CoV-2 pathogenesis
- PMID: 33200027
- PMCID: PMC7657020
- DOI: 10.1016/j.csbj.2020.11.005
In silico and in vitro studies reveal complement system drives coagulation cascade in SARS-CoV-2 pathogenesis
Abstract
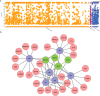
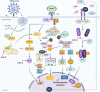
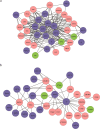
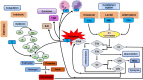
The emergence and continued spread of SARS-CoV-2 have resulted in a public health emergency across the globe. The lack of knowledge on the precise mechanism of viral pathogenesis is impeding medical intervention. In this study, we have taken both in silico and in vitro experimental approaches to unravel the mechanism of viral pathogenesis associated with complement and coagulation pathways. Based on the structural similarities of viral and host proteins, we initially generated a protein-protein interactome profile. Further computational analysis combined with Gene Ontology (GO) analysis and KEGG pathway analysis predicted key annotated pathways associated with viral pathogenesis. These include MAPK signaling, complement, and coagulation cascades, endocytosis, PD-L1 expression, PD-1 checkpoint pathway in cancer and C-type lectin receptor signaling pathways. Degree centrality analysis pinned down to MAPK1, MAPK3, AKT1, and SRC are crucial drivers of signaling pathways and often overlap with the associated pathways. Most strikingly, the complement and coagulation cascade and platelet activation pathways are interconnected, presumably directing thrombotic activity observed in severe or critical cases of COVID-19. This is complemented by in vitro studies of Huh7 cell infection and analysis of the transcriptome and proteomic profile of gene candidates during viral infection. The most known candidates associated with complement and coagulation cascade signaling by KEGG pathway analysis showed significant up-regulated fold change during viral infection. Collectively both in silico and in vitro studies suggest complement and coagulation cascade signaling are a mechanism for intravascular coagulation, thrombotic changes, and associated complications in severe COVID-19 patients.
Keywords: ARDS; Cytokines storm; MAPK1/MAPK3; Neutrophil degranulation; SARS-CoV-2; Thrombosis.
© 2020 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


References
-
- Zhu N.a., Zhang D., Wang W., Li X., Yang B.o., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. - DOI - PMC - PubMed
-
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.-E., Humphrey C.D., Shieh W.-J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.-Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348(20):1953–1966. doi: 10.1056/NEJMoa030781. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
