A Distinct Phylogenetic Cluster of Indian Severe Acute Respiratory Syndrome Coronavirus 2 Isolates
- PMID: 33200080
- PMCID: PMC7543508
- DOI: 10.1093/ofid/ofaa434
A Distinct Phylogenetic Cluster of Indian Severe Acute Respiratory Syndrome Coronavirus 2 Isolates
Abstract
Background: From an isolated epidemic, coronavirus disease 2019 has now emerged as a global pandemic. The availability of genomes in the public domain after the epidemic provides a unique opportunity to understand the evolution and spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus across the globe.
Methods: We performed whole-genome sequencing of 303 Indian isolates, and we analyzed them in the context of publicly available data from India.
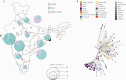
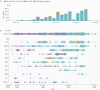
Results: We describe a distinct phylogenetic cluster (Clade I/A3i) of SARS-CoV-2 genomes from India, which encompasses 22% of all genomes deposited in the public domain from India. Globally, approximately 2% of genomes, which to date could not be mapped to any distinct known cluster, fall within this clade.
Conclusions: The cluster is characterized by a core set of 4 genetic variants and has a nucleotide substitution rate of 1.1 × 10-3 variants per site per year, which is lower than the prevalent A2a cluster. Epidemiological assessments suggest that the common ancestor emerged at the end of January 2020 and possibly resulted in an outbreak followed by countrywide spread. To the best of our knowledge, this is the first comprehensive study characterizing this cluster of SARS-CoV-2 in India.
Keywords: COVID-19; Clade I/A3i; India; genetic epidemiology; phylogenomics.
© The Author(s) 2020. Published by Oxford University Press on behalf of Infectious Diseases Society of America.
Figures


References
-
- World Health Organization. Coronavirus disease (COVID-19) Weekly Epidemiological Update. Available at: https://www.who.int/docs/default-source/coronaviruse/situation-reports/2.... Accessed 31 August 2020.
-
- Quick J. NCoV-2019 Sequencing Protocol V2. Protocols.io. 2020; published online March 14 Available at: https://www.protocols.io/view/ncov-2019-sequencing-protocol-v2-bdp7i5rn. Accessed 30 August 2020.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

