Enhanced expression of immune checkpoint receptors during SARS-CoV-2 viral infection
- PMID: 33200082
- PMCID: PMC7658590
- DOI: 10.1016/j.omtm.2020.11.002
Enhanced expression of immune checkpoint receptors during SARS-CoV-2 viral infection
Abstract
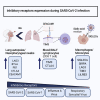
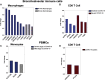
The immune system is tightly regulated by the activity of stimulatory and inhibitory immune receptors. This immune homeostasis is usually disturbed during chronic viral infection. Using publicly available transcriptomic datasets, we conducted in silico analyses to evaluate the expression pattern of 38 selected immune inhibitory receptors (IRs) associated with different myeloid and lymphoid immune cells during coronavirus disease 2019 (COVID-19) infection. Our analyses revealed a pattern of overall upregulation of IR mRNA during severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. A large number of IRs expressed on both lymphoid and myeloid cells were upregulated in nasopharyngeal swabs (NPSs), while lymphoid-associated IRs were specifically upregulated in autopsies, reflecting severe, terminal stage COVID-19 disease. Eight genes (BTLA, LAG3, FCGR2B, PDCD1, CEACAM1, CTLA4, CD72, and SIGLEC7), shared by NPSs and autopsies, were more expressed in autopsies and were directly correlated with viral levels. Single-cell data from blood and bronchoalveolar samples also reflected the observed association between IR upregulation and disease severity. Moreover, compared to SARS-CoV-1, influenza, and respiratory syncytial virus infections, the number and intensities of upregulated IRs were higher in SARS-CoV-2 infections. In conclusion, the immunopathology and severity of COVID-19 could be attributed to dysregulation of different immune inhibitors. Targeting one or more of these immune inhibitors could represent an effective therapeutic approach for the treatment of COVID-19 early and late immune dysregulations.
Keywords: CEACAM1; COVID-19; SARS-CoV-2; SIGLEC10; immune checkpoint inhibitors; immune inhibitory receptors; influenza A virus; lung autopsies; respiratory viral infection; sialic acid.
© 2020 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Upregulation of oxidative stress gene markers during SARS-COV-2 viral infection.Free Radic Biol Med. 2021 Aug 20;172:688-698. doi: 10.1016/j.freeradbiomed.2021.06.018. Epub 2021 Jun 26. Free Radic Biol Med. 2021. PMID: 34186206 Free PMC article.
-
Enhanced Expression of Autoantigens During SARS-CoV-2 Viral Infection.Front Immunol. 2021 Jun 30;12:686462. doi: 10.3389/fimmu.2021.686462. eCollection 2021. Front Immunol. 2021. PMID: 34276672 Free PMC article.
-
GPR183 antagonism reduces macrophage infiltration in influenza and SARS-CoV-2 infection.Eur Respir J. 2023 Mar 9;61(3):2201306. doi: 10.1183/13993003.01306-2022. Print 2023 Mar. Eur Respir J. 2023. PMID: 36396144 Free PMC article.
-
The Role of Immune Regulatory Molecules in COVID-19.Viral Immunol. 2022 Jun;35(5):359-364. doi: 10.1089/vim.2021.0211. Epub 2022 Apr 19. Viral Immunol. 2022. PMID: 35443826 Review.
-
Innate Immune Signaling and Proteolytic Pathways in the Resolution or Exacerbation of SARS-CoV-2 in Covid-19: Key Therapeutic Targets?Front Immunol. 2020 May 28;11:1229. doi: 10.3389/fimmu.2020.01229. eCollection 2020. Front Immunol. 2020. PMID: 32574272 Free PMC article. Review.
Cited by
-
Molecular signature of postmortem lung tissue from COVID-19 patients suggests distinct trajectories driving mortality.Dis Model Mech. 2022 May 1;15(5):dmm049572. doi: 10.1242/dmm.049572. Epub 2022 Jun 6. Dis Model Mech. 2022. PMID: 35438176 Free PMC article.
-
Integrative structural studies of the SARS-CoV-2 spike protein during the fusion process (2022).Curr Res Struct Biol. 2022;4:220-230. doi: 10.1016/j.crstbi.2022.06.004. Epub 2022 Jun 23. Curr Res Struct Biol. 2022. PMID: 35765663 Free PMC article.
-
DeepCoVDR: deep transfer learning with graph transformer and cross-attention for predicting COVID-19 drug response.Bioinformatics. 2023 Jun 30;39(39 Suppl 1):i475-i483. doi: 10.1093/bioinformatics/btad244. Bioinformatics. 2023. PMID: 37387168 Free PMC article.
-
Study of immunological and inflammatory gene response in Indian cohort of COVID- 19 patients by NanoString technology.Immunol Res. 2025 Apr 29;73(1):77. doi: 10.1007/s12026-025-09626-5. Immunol Res. 2025. PMID: 40299133
-
LAG-3 Contribution to T Cell Downmodulation during Acute Respiratory Viral Infections.Viruses. 2023 Jan 3;15(1):147. doi: 10.3390/v15010147. Viruses. 2023. PMID: 36680187 Free PMC article. Review.
References
-
- World Health Organization . 2020. Coronavirus disease (COVID-19): situation report – 205.https://www.who.int/docs/default-source/coronaviruse/situation-reports/2...
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

