C-terminus of Hsp70 Interacting Protein (CHIP) and Neurodegeneration: Lessons from the Bench and Bedside
- PMID: 33200713
- PMCID: PMC8686314
- DOI: 10.2174/1570159X18666201116145507
C-terminus of Hsp70 Interacting Protein (CHIP) and Neurodegeneration: Lessons from the Bench and Bedside
Abstract
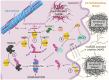
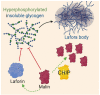
Neurodegenerative diseases are characterized by the increasing dysfunction and death of neurons, resulting in progressive impairment of a person's mobility and/or cognition. Protein misfolding and aggregation are commonly hypothesized to cause neurotoxicity and, eventually, neuronal degeneration that are associated with these diseases. Emerging experimental evidence, as well as recent findings from human studies, reveal that the C-terminus of Hsp70 Interacting Protein (CHIP), or STIP1 Homology and U-box containing Protein 1 (STUB1), is a quality control protein involved in neurodegeneration. Here, we review evidence that CHIP interacts with and plays a role in regulating proteins implicated in the pathogenesis of Parkinson's disease, Alzheimer's disease, amyotrophic lateral sclerosis, and polyglutamine diseases, including Huntington's disease and spinocerebellar ataxias. We also review clinical findings identifying mutations in STUB1 as a cause of both autosomal recessive and autosomal dominant forms of cerebellar ataxia. We propose that CHIP modulation may have therapeutic potential for the treatment of multiple neurodegenerative diseases.
Keywords: Alzheimer's disease; STUB1; STUB1-associated disease.; amyotrophic lateral sclerosis; cerebellar ataxia; huntington disease; neurodegenerative diseases; parkinson's disease; polyglutamine diseases.
Copyright© Bentham Science Publishers; For any queries, please email at epub@benthamscience.net.
Figures




References
-
- Ferri C.P., Prince M., Brayne C., Brodaty H., Fratiglioni L., Ganguli M., Hall K., Hasegawa K., Hendrie H., Huang Y., Jorm A., Mathers C., Menezes P.R., Rimmer E., Scazufca M. Alzheimer’s Disease International. Global Prevalence of Dementia: A Delphi Consensus Study. Lancet Lond. Engl. 2005;366(9503):2112–2117. doi: 10.1016/S0140-6736(05)67889-0. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

