A Two-Armed Probe for In-Cell DEER Measurements on Proteins*
- PMID: 33200852
- PMCID: PMC7839491
- DOI: 10.1002/chem.202002743
A Two-Armed Probe for In-Cell DEER Measurements on Proteins*
Erratum in
-
Corrigendum: A Two-Armed Probe for In-Cell DEER Measurements on Proteins.Chemistry. 2021 Apr 21;27(23):6994. doi: 10.1002/chem.202100954. Epub 2021 Mar 26. Chemistry. 2021. PMID: 33887092 Free PMC article. No abstract available.
Abstract
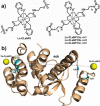
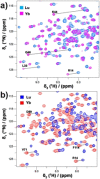
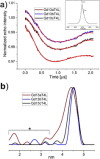
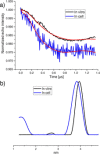
The application of double electron-electron resonance (DEER) with site-directed spin labeling (SDSL) to measure distances in proteins and protein complexes in living cells puts rigorous restraints on the spin-label. The linkage and paramagnetic centers need to resist the reducing conditions of the cell. Rigid attachment of the probe to the protein improves precision of the measured distances. Here, three two-armed GdIII complexes, GdIII -CLaNP13a/b/c were synthesized. Rather than the disulfide linkage of most other CLaNP molecules, a thioether linkage was used to avoid reductive dissociation of the linker. The doubly GdIII labeled N55C/V57C/K147C/T151C variants of T4Lysozyme were measured by 95 GHz DEER. The constructs were measured in vitro, in cell lysate and in Dictyostelium discoideum cells. Measured distances were 4.5 nm, consistent with results from paramagnetic NMR. A narrow distance distribution and typical modulation depth, also in cell, indicate complete and durable labeling and probe rigidity due to the dual attachment sites.
Keywords: EPR spectroscopy; double electron-electron resonance (DEER); gadolinium; protein structures; spin labels.
© 2020 The Authors. Published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Nanometer-scale distance measurements in proteins using Gd3+ spin labeling.J Am Chem Soc. 2010 Jul 7;132(26):9040-8. doi: 10.1021/ja1015662. J Am Chem Soc. 2010. PMID: 20536233
-
Generic tags for Mn(ii) and Gd(iii) spin labels for distance measurements in proteins.Phys Chem Chem Phys. 2017 Oct 11;19(39):26944-26956. doi: 10.1039/c7cp04311b. Phys Chem Chem Phys. 2017. PMID: 28956044
-
EPR Techniques to Probe Insertion and Conformation of Spin-Labeled Proteins in Lipid Bilayers.Methods Mol Biol. 2019;2003:493-528. doi: 10.1007/978-1-4939-9512-7_21. Methods Mol Biol. 2019. PMID: 31218631
-
Characteristics of Gd(III) spin labels for the study of protein conformations.Methods Enzymol. 2021;651:235-290. doi: 10.1016/bs.mie.2021.01.040. Epub 2021 Mar 26. Methods Enzymol. 2021. PMID: 33888206 Review.
-
Elucidating the design principles of photosynthetic electron-transfer proteins by site-directed spin labeling EPR spectroscopy.Biochim Biophys Acta. 2016 May;1857(5):548-556. doi: 10.1016/j.bbabio.2015.08.009. Epub 2015 Sep 1. Biochim Biophys Acta. 2016. PMID: 26334844 Review.
Cited by
-
Is Cys(MTSL) the Best α-Amino Acid Residue to Electron Spin Labeling of Synthetically Accessible Peptide Molecules with Nitroxides?ACS Omega. 2022 Jan 31;7(6):5154-5165. doi: 10.1021/acsomega.1c06227. eCollection 2022 Feb 15. ACS Omega. 2022. PMID: 35187331 Free PMC article.
-
Flavoproteins as native and genetically encoded spin probes for in cell ESR spectroscopy.Nat Commun. 2025 Jul 1;16(1):5406. doi: 10.1038/s41467-025-60623-6. Nat Commun. 2025. PMID: 40595551 Free PMC article.
-
Paramagnetic Chemical Probes for Studying Biological Macromolecules.Chem Rev. 2022 May 25;122(10):9571-9642. doi: 10.1021/acs.chemrev.1c00708. Epub 2022 Jan 27. Chem Rev. 2022. PMID: 35084831 Free PMC article. Review.
-
Dance with spins: site-directed spin labeling coupled to electron paramagnetic resonance spectroscopy directly inside cells.Chem Commun (Camb). 2023 Jan 31;59(10):1274-1284. doi: 10.1039/d2cc05907j. Chem Commun (Camb). 2023. PMID: 36633152 Free PMC article. Review.
References
-
- Schiemann O., Prisner T. F., Q. Rev. Biophys. 2007, 40, 1–53. - PubMed
-
- Jeschke G., Polyhach Y., Phys. Chem. Chem. Phys. 2007, 9, 1895–1910. - PubMed
-
- Borbat P. P., Freed J. H., in: Structural Information from Spin-Labels and Intrinsic Paramagnetic, Centres in the Biosciences (Eds.: R. Timmel C., R. Harmer J.), Springer, Heidelberg, 2013, pp. 1–82.
MeSH terms
Substances
Grants and funding
- CSC grant to Q. M, No. 201506870013/Chinese Scholarship Council
- Grant No. 711.014.003/Nederlandse Organisatie voor Wetenschappelijk Onderzoek
- Spinoza Prijs 2014 for Prof. Dirk Bouwmeester/Nederlandse Organisatie voor Wetenschappelijk Onderzoek
- Fraunhofer Attract grant "3DNanoCell"/Fraunhofer Society
- Fraunhofer ICON grant "BioSensing"/Fraunhofer Society
LinkOut - more resources
Full Text Sources

