STING induces LC3B lipidation onto single-membrane vesicles via the V-ATPase and ATG16L1-WD40 domain
- PMID: 33201170
- PMCID: PMC7716379
- DOI: 10.1083/jcb.202009128
STING induces LC3B lipidation onto single-membrane vesicles via the V-ATPase and ATG16L1-WD40 domain
Abstract
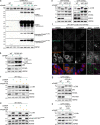
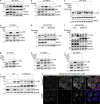
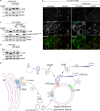
Following the detection of cytosolic double-stranded DNA from viral or bacterial infection in mammalian cells, cyclic dinucleotide activation of STING induces interferon β expression to initiate innate immune defenses. STING activation also induces LC3B lipidation, a classical but equivocal marker of autophagy, that promotes a cell-autonomous antiviral response that arose before evolution of the interferon pathway. We report that STING activation induces LC3B lipidation onto single-membrane perinuclear vesicles mediated by ATG16L1 via its WD40 domain, bypassing the requirement of canonical upstream autophagy machinery. This process is blocked by bafilomycin A1 that binds and inhibits the vacuolar ATPase (V-ATPase) and by SopF, a bacterial effector that catalytically modifies the V-ATPase to inhibit LC3B lipidation via ATG16L1. These results indicate that activation of the cGAS-STING pathway induces V-ATPase-dependent LC3B lipidation that may mediate cell-autonomous host defense, an unanticipated mechanism that is distinct from LC3B lipidation onto double-membrane autophagosomes.
© 2020 Fischer et al. This article is available under a Creative Commons License (Attribution 4.0 International, as described at https://creativecommons.org/licenses/by/4.0/).
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

