Congenital heart disease risk loci identified by genome-wide association study in European patients
- PMID: 33201861
- PMCID: PMC7810487
- DOI: 10.1172/JCI141837
Congenital heart disease risk loci identified by genome-wide association study in European patients
Abstract
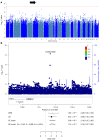
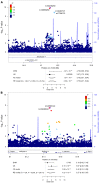
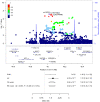
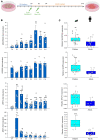
Genetic factors undoubtedly affect the development of congenital heart disease (CHD) but still remain ill defined. We sought to identify genetic risk factors associated with CHD and to accomplish a functional analysis of SNP-carrying genes. We performed a genome-wide association study (GWAS) of 4034 White patients with CHD and 8486 healthy controls. One SNP on chromosome 5q22.2 reached genome-wide significance across all CHD phenotypes and was also indicative for septal defects. One region on chromosome 20p12.1 pointing to the MACROD2 locus identified 4 highly significant SNPs in patients with transposition of the great arteries (TGA). Three highly significant risk variants on chromosome 17q21.32 within the GOSR2 locus were detected in patients with anomalies of thoracic arteries and veins (ATAV). Genetic variants associated with ATAV are suggested to influence the expression of WNT3, and the variant rs870142 related to septal defects is proposed to influence the expression of MSX1. We analyzed the expression of all 4 genes during cardiac differentiation of human and murine induced pluripotent stem cells in vitro and by single-cell RNA-Seq analyses of developing murine and human hearts. Our data show that MACROD2, GOSR2, WNT3, and MSX1 play an essential functional role in heart development at the embryonic and newborn stages.
Keywords: Cardiology; Cardiovascular disease; Genetics; Molecular genetics; iPS cells.
Conflict of interest statement
Figures



Comment in
-
Unraveling the genomic basis of congenital heart disease.J Clin Invest. 2021 Jan 19;131(2):e145377. doi: 10.1172/JCI145377. J Clin Invest. 2021. PMID: 33463545 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

