Unique Hydrogen Bonding of Adenine with the Oxidatively Damaged Base 8-Oxoguanine Enables Specific Recognition and Repair by DNA Glycosylase MutY
- PMID: 33202125
- PMCID: PMC9187209
- DOI: 10.1021/jacs.0c06767
Unique Hydrogen Bonding of Adenine with the Oxidatively Damaged Base 8-Oxoguanine Enables Specific Recognition and Repair by DNA Glycosylase MutY
Abstract
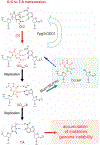
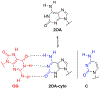
The DNA glycosylase MutY prevents deleterious mutations resulting from guanine oxidation by recognition and removal of adenine (A) misincorporated opposite 8-oxo-7,8-dihydroguanine (OG). Correct identification of OG:A is crucial to prevent improper and detrimental MutY-mediatedadenine excision from G:A or T:A base pairs. Here we present a structure-activity relationship (SAR) study using analogues of A to probe the basis for OG:A specificity of MutY. We correlate observed in vitro MutY activity on A analogue substrates with their experimental and calculated acidities to provide mechanistic insight into the factors influencing MutY base excision efficiency. These data show that H-bonding and electrostatic interactions of the base within the MutY active site modulate the lability of the N-glycosidic bond. A analogues that were not excised from duplex DNA as efficiently as predicted by calculations provided insight into other required structural features, such as steric fit and H-bonding within the active site for proper alignment with MutY catalytic residues. We also determined MutY-mediated repair of A analogues paired with OG within the context of a DNA plasmid in bacteria. Remarkably, the magnitudes of decreased in vitro MutY excision rates with different A analogue duplexes do not correlate with the impact on overall MutY-mediated repair. The feature that most strongly correlated with facile cellular repair was the ability of the A analogues to H-bond with the Hoogsteen face of OG. Notably, base pairing of A with OG uniquely positions the 2-amino group of OG in the major groove and provides a means to indirectly select only these inappropriately placed adenines for excision. This highlights the importance of OG lesion detection for efficient MutY-mediated cellular repair. The A analogue SARs also highlight the types of modifications tolerated by MutY and will guide the development of specific probes and inhibitors of MutY.
Figures




References
-
- Banda DM; Nuñez NN; Burnside MA; Bradshaw KM; David SS Repair of 8-OxoG:A Mismatches by the MUTYH Glycosylase: Mechanism, Metals and Medicine. Free Radic. Biol. Med 2017, 107, 202–215. https://doi.org/10.1016/j.freeradbiomed.2017.01.008. - DOI - PMC - PubMed
-
- Drabløs F; Feyzi E; Aas PA; Vaagbø CB; Kavli B; Bratlie MS; Peña-Diaz J; Otterlei M; Slupphaug G; Krokan HE Alkylation Damage in DNA and RNA—Repair Mechanisms and Medical Significance. DNA Repair (Amst). 2004, 3 (11), 1389–1407. https://doi.org/10.1016/j.dnarep.2004.05.004. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

