Multiplexed Functional Assessment of Genetic Variants in CARD11
- PMID: 33202260
- PMCID: PMC7820631
- DOI: 10.1016/j.ajhg.2020.10.015
Multiplexed Functional Assessment of Genetic Variants in CARD11
Abstract
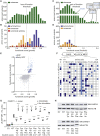
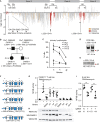
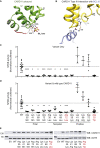
Genetic testing has increased the number of variants identified in disease genes, but the diagnostic utility is limited by lack of understanding variant function. CARD11 encodes an adaptor protein that expresses dominant-negative and gain-of-function variants associated with distinct immunodeficiencies. Here, we used a "cloning-free" saturation genome editing approach in a diploid cell line to simultaneously score 2,542 variants for decreased or increased function in the region of CARD11 associated with immunodeficiency. We also described an exon-skipping mechanism for CARD11 dominant-negative activity. The classification of reported clinical variants was sensitive (94.6%) and specific (88.9%), which rendered the data immediately useful for interpretation of seven coding and splicing variants implicated in immunodeficiency found in our clinic. This approach is generalizable for variant interpretation in many other clinically actionable genes, in any relevant cell type.
Keywords: B cells; CARD11; gene editing; immune dysregulation; lymphoma; primary immune deficiency; saturation genome editing; variant interpretation.
Copyright © 2020 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

