NEAT1/miR-200b-3p/SMAD2 axis promotes progression of melanoma
- PMID: 33202380
- PMCID: PMC7746346
- DOI: 10.18632/aging.103909
NEAT1/miR-200b-3p/SMAD2 axis promotes progression of melanoma
Abstract
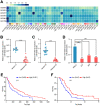
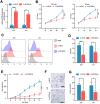
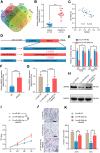
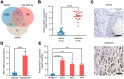
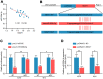
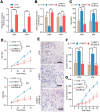
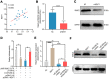
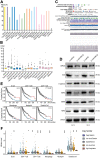
Melanoma is a skin malignancy with a high mutation frequency of genetic alterations. MicroRNA (miR)-200b-3p is involved in various cancers, while in melanoma its bio-function remains unknown. In this study, we found that miR-200b-3p was down-regulated in melanoma tissues and cell lines compared to benign nevus cells. Overexpression of miR-200b-3p significantly inhibited the proliferation and invasion of melanoma cells. According to bioinformatics analysis and sequencing data, we supposed that SMAD family member 2 (SMAD2) was the target gene and nuclear enriched abundant transcript 1 (NEAT1) was the upstream long non-coding RNA (lncRNA) of miR-200b-3p. These predictions were verified by western blotting and quantitative real-time reverse transcription PCR (RT-qPCR). Luciferase reporter assays revealed that NEAT1 up-regulated SMAD2 by directly sponging miR-200b-3p. In vitro and in vivo, we demonstrated that both NEAT1 and SMAD2 could promote the proliferation and invasion of melanoma cells, and these effects were reversed by up-regulating miR-200b-3p. In addition, NEAT1/miR-200b-3p/SMAD2 axis promoted melanoma progression by activating EMT signaling pathway and immune responses. Taken together, the NEAT1/miR-200b-3p/SMAD2 signaling pathway promotes melanoma via activation of EMT, cell invasion and is related with immune responses, which provides new insights into the molecular mechanisms and therapeutic targets for melanoma.
Keywords: EMT; NEAT1; SMAD2; melanoma; miR-200b-3p.
Conflict of interest statement
Figures
References
-
- Sundstrøm T, Espedal H, Harter PN, Fasmer KE, Skaftnesmo KO, Horn S, Hodneland E, Mittelbronn M, Weide B, Beschorner R, Bender B, Rygh CB, Lund-Johansen M, et al.. Melanoma brain metastasis is independent of lactate dehydrogenase a expression. Neuro Oncol. 2015; 17:1374–85. 10.1093/neuonc/nov040 - DOI - PMC - PubMed
-
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Børresen-Dale AL, Boyault S, Burkhardt B, Butler AP, et al., and Australian Pancreatic Cancer Genome Initiative, and ICGC Breast Cancer Consortium, and ICGC MMML-Seq Consortium, and ICGC PedBrain. Signatures of mutational processes in human cancer. Nature. 2013; 500:415–21. 10.1038/nature12477 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

