Genetic and Antigenic Evolution of European Swine Influenza A Viruses of HA-1C (Avian-Like) and HA-1B (Human-Like) Lineages in France from 2000 to 2018
- PMID: 33202972
- PMCID: PMC7697621
- DOI: 10.3390/v12111304
Genetic and Antigenic Evolution of European Swine Influenza A Viruses of HA-1C (Avian-Like) and HA-1B (Human-Like) Lineages in France from 2000 to 2018
Abstract
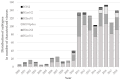
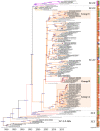
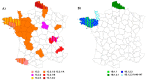
This study evaluated the genetic and antigenic evolution of swine influenza A viruses (swIAV) of the two main enzootic H1 lineages, i.e., HA-1C (H1av) and -1B (H1hu), circulating in France between 2000 and 2018. SwIAV RNAs extracted from 1220 swine nasal swabs were hemagglutinin/neuraminidase (HA/NA) subtyped by RT-qPCRs, and 293 virus isolates were sequenced. In addition, 146 H1avNy and 105 H1huNy strains were submitted to hemagglutination inhibition tests. H1avN1 (66.5%) and H1huN2 (25.4%) subtypes were predominant. Most H1 strains belonged to HA-1C.2.1 or -1B.1.2.3 clades, but HA-1C.2, -1C.2.2, -1C.2.3, -1B.1.1, and -1B.1.2.1 clades were also detected sporadically. Within HA-1B.1.2.3 clade, a group of strains named "Δ146-147" harbored several amino acid mutations and a double deletion in HA, that led to a marked antigenic drift. Phylogenetic analyses revealed that internal segments belonged mainly to the "Eurasian avian-like lineage", with two distinct genogroups for the M segment. In total, 17 distinct genotypes were identified within the study period. Reassortments of H1av/H1hu strains with H1N1pdm virus were rarely evidenced until 2018. Analysis of amino acid sequences predicted a variability in length of PB1-F2 and PA-X proteins and identified the appearance of several mutations in PB1, PB1-F2, PA, NP and NS1 proteins that could be linked to virulence, while markers for antiviral resistance were identified in N1 and N2. Altogether, diversity and evolution of swIAV recall the importance of disrupting the spreading of swIAV within and between pig herds, as well as IAV inter-species transmissions.
Keywords: Eurasian avian-like lineage; H1N1; H1N2; antigenic drift; genetic diversity; genotype; matrix protein; surveillance; swine influenza; virus evolution.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

