Histological Evaluation and Gene Expression Profiling of Autophagy-Related Genes for Cartilage of Young and Senescent Rats
- PMID: 33203108
- PMCID: PMC7697851
- DOI: 10.3390/ijms21228607
Histological Evaluation and Gene Expression Profiling of Autophagy-Related Genes for Cartilage of Young and Senescent Rats
Abstract
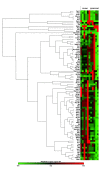
Autophagy is a cellular mechanism that protects cells from stress by digesting non-functional cellular components. In the cartilage, chondrocytes depend on autophagy as a principal mechanism to maintain cellular homeostasis. This protective role diminishes prior to the structural damage that normally occurs during aging. Considering that aging is the main risk factor for osteoarthritis, evaluating the expression of genes associated with autophagy in senescent cartilage might allow for the identification of potential therapeutic targets for treatment. Thus, we studied two groups of young and senescent rats. A histological analysis of cartilage and gene expression quantification for autophagy-related genes were performed. In aged cartilage, morphological changes were observed, such as an increase in cartilage degeneration as measured by the modified Mankin score, a decrease in the number of chondrocytes and collagen II (Col2a1), and an increase in matrix metalloproteinase 13 (Mmp13). Moreover, 84 genes associated with autophagy were evaluated by a PCR array analysis, and 15 of them were found to be significantly decreased with aging. Furthermore, an in silico analysis based on by two different bioinformatics software tools revealed that several processes including cellular homeostasis, autophagosome assembly, and aging-as well as several biological pathways such as autophagy, insulin-like growth factor 1 (IGF-1) signaling, PI3K (phosphoinositide 3-kinase)/AKT (serine/threonine kinase) signaling, and mammalian target of rapamycin (mTOR) signaling-were enriched. In conclusion, the analysis identified some potential targets for osteoarthritis treatment that would allow for the development of new therapeutic strategies for this chronic disease.
Keywords: aging; autophagy; osteoarthritis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Nabavi S.F., Sureda A., Dehpour A.R., Shirooie S., Silva A.S., Devi K.P., Ahmed T., Ishaq N., Hashim R., Sobarzo-Sanchez E., et al. Regulation of autophagy by polyphenols: Paving the road for treatment of neurodegeneration. Biotechnol. Adv. 2018;36:1768–1778. doi: 10.1016/j.biotechadv.2017.12.001. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

