Sensing the allosteric force
- PMID: 33203849
- PMCID: PMC7673989
- DOI: 10.1038/s41467-020-19689-7
Sensing the allosteric force
Abstract
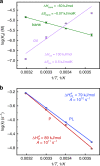
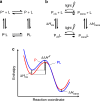
Allosteric regulation is an innate control in most metabolic and signalling cascades that enables living organisms to adapt to the changing environment by tuning the affinity and regulating the activity of target proteins. For a microscopic understanding of this process, a protein system has been designed in such a way that allosteric communication between the binding and allosteric site can be observed in both directions. To that end, an azobenzene-derived photoswitch has been linked to the α3-helix of the PDZ3 domain, arguably the smallest allosteric protein with a clearly identifiable binding and allosteric site. Photo-induced trans-to-cis isomerisation of the photoswitch increases the binding affinity of a small peptide ligand to the protein up to 120-fold, depending on temperature. At the same time, ligand binding speeds up the thermal cis-to-trans back-isomerisation rate of the photoswitch. Based on the energetics of the four states of the system (cis vs trans and ligand-bound vs free), the concept of an allosteric force is introduced, which can be used to drive chemical reactions.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

