Distinct oligodendrocyte populations have spatial preference and different responses to spinal cord injury
- PMID: 33203872
- PMCID: PMC7673029
- DOI: 10.1038/s41467-020-19453-x
Distinct oligodendrocyte populations have spatial preference and different responses to spinal cord injury
Abstract
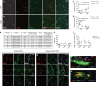
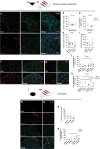
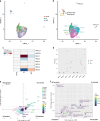
Mature oligodendrocytes (MOLs) show transcriptional heterogeneity, the functional consequences of which are unclear. MOL heterogeneity might correlate with the local environment or their interactions with different neuron types. Here, we show that distinct MOL populations have spatial preference in the mammalian central nervous system (CNS). We found that MOL type 2 (MOL2) is enriched in the spinal cord when compared to the brain, while MOL types 5 and 6 (MOL5/6) increase their contribution to the OL lineage with age in all analyzed regions. MOL2 and MOL5/6 also have distinct spatial preference in the spinal cord regions where motor and sensory tracts run. OL progenitor cells (OPCs) are not specified into distinct MOL populations during development, excluding a major contribution of OPC intrinsic mechanisms determining MOL heterogeneity. In disease, MOL2 and MOL5/6 present different susceptibility during the chronic phase following traumatic spinal cord injury. Our results demonstrate that the distinct MOL populations have different spatial preference and different responses to disease.
Conflict of interest statement
M.N. and M.M.H. held shares in Cartana AB, a company commercializing in situ sequencing reagents. E.M.F. is currently employed as an Associate Editor of
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

