XPC deficiency increases risk of hematologic malignancies through mutator phenotype and characteristic mutational signature
- PMID: 33203900
- PMCID: PMC7672101
- DOI: 10.1038/s41467-020-19633-9
XPC deficiency increases risk of hematologic malignancies through mutator phenotype and characteristic mutational signature
Abstract
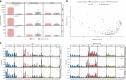
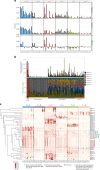
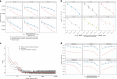
Recent studies demonstrated a dramatically increased risk of leukemia in patients with a rare genetic disorder, Xeroderma Pigmentosum group C (XP-C), characterized by constitutive deficiency of global genome nucleotide excision repair (GG-NER). The genetic mechanisms of non-skin cancers in XP-C patients remain unexplored. In this study, we analyze a unique collection of internal XP-C tumor genomes including 6 leukemias and 2 sarcomas. We observe a specific mutational pattern and an average of 25-fold increase of mutation rates in XP-C versus sporadic leukemia which we presume leads to its elevated incidence and early appearance. We describe a strong mutational asymmetry with respect to transcription and the direction of replication in XP-C tumors suggesting association of mutagenesis with bulky purine DNA lesions of probably endogenous origin. These findings suggest existence of a balance between formation and repair of bulky DNA lesions by GG-NER in human body cells which is disrupted in XP-C patients.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Kraemer, K. H., Lee, M. M., Andrews, A. D. & Lambert, W. C. The role of sunlight and DNA repair in melanoma and nonmelanoma skin cancer: the Xeroderma Pigmentosum paradigm. Arch. Dermatol. 10.1001/archderm.1994.01690080084012 (1994). - PubMed
-
- Kraemer, K. H. Xeroderma pigmentosum. Cutaneous, ocular, and neurologic abnormalities in 830 published cases. Arch. Dermatol.10.1136/jmg.2010.083022 (1987). - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources

