Ecological drivers of genetic connectivity for African malaria vectors Anopheles gambiae and An. arabiensis
- PMID: 33203917
- PMCID: PMC7673128
- DOI: 10.1038/s41598-020-76248-2
Ecological drivers of genetic connectivity for African malaria vectors Anopheles gambiae and An. arabiensis
Abstract
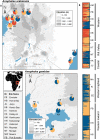
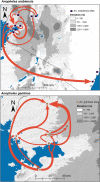
Anopheles gambiae and An. arabiensis are major malaria vectors in sub-Saharan Africa. Knowledge of how geographical factors drive the dispersal and gene flow of malaria vectors can help in combatting insecticide resistance spread and planning new vector control interventions. Here, we used a landscape genetics approach to investigate population relatedness and genetic connectivity of An. gambiae and An. arabiensis across Kenya and determined the changes in mosquito population genetic diversity after 20 years of intensive malaria control efforts. We found a significant reduction in genetic diversity in An. gambiae, but not in An. arabiensis as compared to prior to the 20-year period in western Kenya. Significant population structure among populations was found for both species. The most important ecological driver for dispersal and gene flow of An. gambiae and An. arabiensis was tree cover and cropland, respectively. These findings highlight that human induced environmental modifications may enhance genetic connectivity of malaria vectors.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- World Health Organization . World malaria report 2019. Geneva: WHO; 2019.
-
- Wirtz RA, Burkot TR. Detection of malarial parasites in mosquitoes. In: Maudlin I, Sinha RC, editors. Advances in Disease Vector Research. New York: Sprinter; 1991.
-
- Macdonald G. The Epidemiology and Control of Malaria. London: Oxford Univ. Press; 1957.

