YTHDF1-enhanced iron metabolism depends on TFRC m6A methylation
- PMID: 33204330
- PMCID: PMC7667694
- DOI: 10.7150/thno.51231
YTHDF1-enhanced iron metabolism depends on TFRC m6A methylation
Abstract
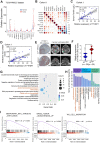
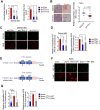
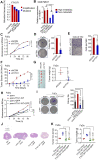
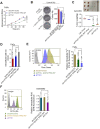
Background: Among head and neck squamous cell carcinomas (HNSCCs), hypopharyngeal squamous cell carcinoma (HPSCC) has the worst prognosis. Iron metabolism, which plays a crucial role in tumor progression, is mainly regulated by alterations to genes and post-transcriptional processes. The recent discovery of the N6-methyladenosine (m6A) modification has expanded the realm of previously undiscovered post-transcriptional gene regulation mechanisms in eukaryotes. Many studies have demonstrated that m6A methylation represents a distinct layer of epigenetic deregulation in carcinogenesis and tumor proliferation. However, the status of m6A modification and iron metabolism in HPSCC remains unknown. Methods: Bioinformatics analysis, sample analysis, and transcriptome sequencing were performed to evaluate the correlation between m6A modification and iron metabolism. Iron metabolic and cell biological analyses were conducted to evaluate the effect of the m6A reader YTHDF1 on HPSCC proliferation and iron metabolism. Transcriptome-wide m6A-seq and RIP-seq data were mapped to explore the molecular mechanism of YTHDF1 function in HPSCC. Results: YTHDF1 was found to be closely associated with ferritin levels and intratumoral iron concentrations in HPSCC patients at Sir Run Run Shaw Hospital. YTHDF1 induced-HPSCC tumorigenesis depends on iron metabolism in vivo in vitro. Mechanistically, YTHDF1 methyltransferase domain interacts with the 3'UTR and 5'UTR of TRFC mRNA, then further positively regulates translation of m6A-modified TFRC mRNA. Gain-of-function and loss-of-function analyses validated the finding showing that TFRC is a crucial target gene for YTHDF1-mediated increases in iron metabolism. Conclusion: YTHDF1 enhanced TFRC expression in HPSCC through an m6A-dependent mechanism. From a therapeutic perspective, targeting YTHDF1 and TFRC-mediated iron metabolism may be a promising strategy for HPSCC.
Keywords: Hypopharyngeal squamous cell carcinoma; Iron metabolism; N6-methyladenosine (m6A) modification; TFRC; YTHDF1.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures



References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. - PubMed
-
- Gatta G, Botta L, Sanchez MJ, Anderson LA, Pierannunzio D, Licitra L. Prognoses and improvement for head and neck cancers diagnosed in Europe in early 2000s: The EUROCARE-5 population-based study. Eur J Cancer. 2015;51:2130–43. - PubMed
-
- Carvalho AL, Nishimoto IN, Califano JA, Kowalski LP. Trends in incidence and prognosis for head and neck cancer in the United States: a site-specific analysis of the SEER database. Int J Cancer. 2005;114:806–16. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

