Single-Cell Profiling Reveals Divergent, Globally Patterned Immune Responses in Murine Skin Inflammation
- PMID: 33205009
- PMCID: PMC7648132
- DOI: 10.1016/j.isci.2020.101582
Single-Cell Profiling Reveals Divergent, Globally Patterned Immune Responses in Murine Skin Inflammation
Abstract
Inflammatory response heterogeneity has impeded high-resolution dissection of diverse immune cell populations during activation. We characterize mouse cutaneous immune cells by single-cell RNA sequencing, after inducing inflammation using imiquimod and oxazolone dermatitis models. We identify 13 CD45+ subpopulations, which broadly represent most functionally characterized immune cell types. Oxazolone pervasively upregulates Jak2/Stat3 expression across T cells and antigen-presenting cells (APCs). Oxazolone also induces Il4/Il13 expression in newly infiltrating basophils, and Il4ra and Ccl24, most prominently in APCs. In contrast, imiquimod broadly upregulates Il17/Il22 and Ccl4/Ccl5. A comparative analysis of single-cell inflammatory transcriptional responses reveals that APC response to oxazolone is tightly restricted by cell identity, whereas imiquimod enforces shared programs on multiple APC populations in parallel. These global molecular patterns not only contrast immune responses on a systems level but also suggest that the mechanisms of new sources of inflammation can eventually be deduced by comparison to known signatures.
Keywords: Immunology; Systems Biology.
Conflict of interest statement
A.J.S. and S.C.B. are employees of ImmunityBio Inc, S.C.B. is an equity holder of ImmunityBio Inc. The remaining authors disclose no conflicts.
Figures



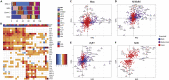
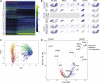

References
-
- Akahoshi M., Song C.H., Piliponsky A.M., Metz M., Guzzetta A., Abrink M., Schlenner S.M., Feyerabend T.B., Rodewald H.-R., Pejler G. Mast cell chymase reduces the toxicity of Gila monster venom, scorpion venom, and vasoactive intestinal polypeptide in mice. J. Clin. Invest. 2011;121:4180–4191. - PMC - PubMed
-
- Bros M., Dexheimer N., Ross R., Trojandt S., Höhn Y., Tampe J., Sutter A., Jährling F., Grabbe S., Reske-Kunz A.B. Differential gene expression analysis identifies murine Cacnb3 as strongly upregulated in distinct dendritic cell populations upon stimulation. Gene. 2011;472:18–27. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

