Spectral Jaccard Similarity: A New Approach to Estimating Pairwise Sequence Alignments
- PMID: 33205128
- PMCID: PMC7660437
- DOI: 10.1016/j.patter.2020.100081
Spectral Jaccard Similarity: A New Approach to Estimating Pairwise Sequence Alignments
Abstract
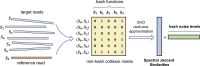
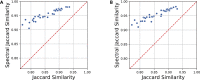
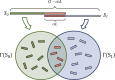
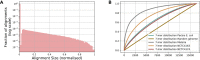
Pairwise sequence alignment is often a computational bottleneck in genomic analysis pipelines, particularly in the context of third-generation sequencing technologies. To speed up this process, the pairwise k-mer Jaccard similarity is sometimes used as a proxy for alignment size in order to filter pairs of reads, and min-hashes are employed to efficiently estimate these similarities. However, when the k-mer distribution of a dataset is significantly non-uniform (e.g., due to GC biases and repeats), Jaccard similarity is no longer a good proxy for alignment size. In this work, we introduce a min-hash-based approach for estimating alignment sizes called Spectral Jaccard Similarity, which naturally accounts for uneven k-mer distributions. The Spectral Jaccard Similarity is computed by performing a singular value decomposition on a min-hash collision matrix. We empirically show that this new metric provides significantly better estimates for alignment sizes, and we provide a computationally efficient estimator for these spectral similarity scores.
Keywords: DNA sequencing; Jaccard similarity; genome assembly; locality-sensitive hashing; min-hash; sequence alignment; singular value decomposition; spectral methods.
© 2020 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures


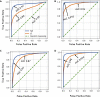
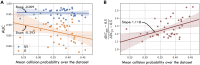

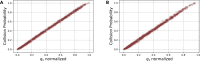
References
-
- Weirather Jason L., de Cesare Mariateresa, Wang Yunhao, Piazza Paolo, Sebastiano Vittorio, Wang Xiu-Jie, Buck David, Au Kin Fai. Comprehensive comparison of pacific biosciences and oxford nanopore technologies and their applications to transcriptome analysis. F1000Res. 2017;6 doi: 10.12688/f1000research.10571.2. - DOI - PMC - PubMed
-
- Needleman Saul B., Wunsch Christian D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970;48:443–453. - PubMed
-
- Smith Temple F., Waterman Michael S. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–197. - PubMed
-
- Myers Eugene W. An O(nd) difference algorithm and its variations. Algorithmica. 1986;1:251–266.
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

