Stability of SARS-CoV-2 phylogenies
- PMID: 33206635
- PMCID: PMC7721162
- DOI: 10.1371/journal.pgen.1009175
Stability of SARS-CoV-2 phylogenies
Abstract
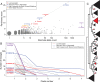
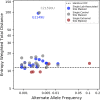
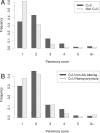
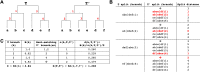
The SARS-CoV-2 pandemic has led to unprecedented, nearly real-time genetic tracing due to the rapid community sequencing response. Researchers immediately leveraged these data to infer the evolutionary relationships among viral samples and to study key biological questions, including whether host viral genome editing and recombination are features of SARS-CoV-2 evolution. This global sequencing effort is inherently decentralized and must rely on data collected by many labs using a wide variety of molecular and bioinformatic techniques. There is thus a strong possibility that systematic errors associated with lab-or protocol-specific practices affect some sequences in the repositories. We find that some recurrent mutations in reported SARS-CoV-2 genome sequences have been observed predominantly or exclusively by single labs, co-localize with commonly used primer binding sites and are more likely to affect the protein-coding sequences than other similarly recurrent mutations. We show that their inclusion can affect phylogenetic inference on scales relevant to local lineage tracing, and make it appear as though there has been an excess of recurrent mutation or recombination among viral lineages. We suggest how samples can be screened and problematic variants removed, and we plan to regularly inform the scientific community with our updated results as more SARS-CoV-2 genome sequences are shared (https://virological.org/t/issues-with-sars-cov-2-sequencing-data/473 and https://virological.org/t/masking-strategies-for-sars-cov-2-alignments/480). We also develop tools for comparing and visualizing differences among very large phylogenies and we show that consistent clade- and tree-based comparisons can be made between phylogenies produced by different groups. These will facilitate evolutionary inferences and comparisons among phylogenies produced for a wide array of purposes. Building on the SARS-CoV-2 Genome Browser at UCSC, we present a toolkit to compare, analyze and combine SARS-CoV-2 phylogenies, find and remove potential sequencing errors and establish a widely shared, stable clade structure for a more accurate scientific inference and discourse.
Conflict of interest statement
Conflict of interest statement. A.S.H. and D.H. receive royalties from the sale of UCSC Genome Browser source code, LiftOver, GBiB, and GBiC licenses to commercial entities. RL works as an advisor to GISAID.
Figures







References
-
- NCBI Staff. NCBI Insights: INSDC Statement on SARS-CoV-2 sequence data sharing during COVID-19. 17 Aug 2020 [cited 26 Aug 2020]. Available: https://ncbiinsights.ncbi.nlm.nih.gov/2020/08/17/insdc-covid-data-sharing/
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

