Curcumin Treatment Identifies Therapeutic Targets within Biomarkers of Liver Colonization by Highly Invasive Mesothelioma Cells-Potential Links with Sarcomas
- PMID: 33207594
- PMCID: PMC7696465
- DOI: 10.3390/cancers12113384
Curcumin Treatment Identifies Therapeutic Targets within Biomarkers of Liver Colonization by Highly Invasive Mesothelioma Cells-Potential Links with Sarcomas
Abstract
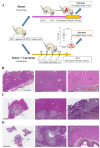
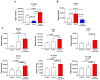
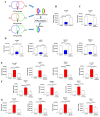
Investigations of liver metastatic colonization suggest that the microenvironment is preordained to be intrinsically hospitable to the invasive cancer cells. To identify molecular determinants of that organotropism and potential therapeutic targets, we conducted proteomic analyses of the liver in an aggressive model of sarcomatoid peritoneal mesothelioma (M5-T1). The quantitative changes between SWATH-MS (sequential window acquisition of all theoretical fragmentation spectra) proteotype patterns of the liver from normal rats (G1), adjacent non-tumorous liver from untreated tumor-bearing rats (G2), and liver from curcumin-treated rats without hepatic metastases (G3) were compared. The results identified 12 biomarkers of raised immune response against M5-T1 cells in G3 and 179 liver biomarker changes in (G2 vs. G1) and (G3 vs. G2) but not in (G3 vs. G1). Cross-comparing these 179 candidates with proteins showing abundance changes related to increasing invasiveness in four different rat mesothelioma tumor models identified seven biomarkers specific to the M5-T1 tumor. Finally, analysis of correlations between these seven biomarkers, purine nucleoside phosphorylase being the main biomarker of immune response, and the 179 previously identified proteins revealed a network orchestrating liver colonization and treatment efficacy. These results highlight the links between potential targets, raising interesting prospects for optimizing therapies against highly invasive cancer cells exhibiting a sarcomatoid phenotype and sarcoma cells.
Keywords: biomarkers; curcumin; invasiveness; liver colonization; malignant mesothelioma; metastatic tumor cells; proteomics; sarcomatoid phenotype.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
Grants and funding
LinkOut - more resources
Full Text Sources

