Dysfunctional Mitochondrial Dynamic and Oxidative Phosphorylation Precedes Cardiac Dysfunction in R120G-αB-Crystallin-Induced Desmin-Related Cardiomyopathy
- PMID: 33208022
- PMCID: PMC7763772
- DOI: 10.1161/JAHA.120.017195
Dysfunctional Mitochondrial Dynamic and Oxidative Phosphorylation Precedes Cardiac Dysfunction in R120G-αB-Crystallin-Induced Desmin-Related Cardiomyopathy
Abstract
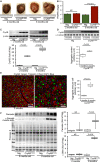
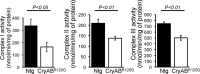
Background The mutated α-B-Crystallin (CryABR120G) mouse model of desmin-related myopathy (DRM) shows an age-dependent onset of pathologic cardiac remodeling and progression of heart failure. CryABR120G expression in cardiomyocytes affects the mitochondrial spatial organization within the myofibrils, but the molecular perturbation within the mitochondria in the relation of the overall course of the proteotoxic disease remains unclear. Methods and Results CryABR120G mice show an accumulation of electron-dense aggregates and myofibrillar degeneration associated with the development of cardiac dysfunction. Though extensive studies demonstrated that these altered ultrastructural changes cause cardiac contractility impairment, the molecular mechanism of cardiomyocyte death remains elusive. Here, we explore early pathological processes within the mitochondria contributing to the contractile dysfunction and determine the pathogenic basis for the heart failure observed in the CryABR120G mice. In the present study, we report that the CryABR120G mice transgenic hearts undergo altered mitochondrial dynamics associated with increased level of dynamin-related protein 1 and decreased level of optic atrophy type 1 as well as mitofusin 1 over the disease process. In association with these changes, an altered level of the components of mitochondrial oxidative phosphorylation and pyruvate dehydrogenase complex regulatory proteins occurs before the manifestation of pathologic adverse remodeling in the CryABR120G hearts. Mitochondria isolated from CryABR120G transgenic hearts without visible pathology show decreased electron transport chain complex activities and mitochondrial respiration. Taken together, we demonstrated the involvement of mitochondria in the pathologic remodeling and progression of DRM-associated cellular dysfunction. Conclusions Mitochondrial dysfunction in the form of altered mitochondrial dynamics, oxidative phosphorylation and pyruvate dehydrogenase complex proteins level, abnormal electron transport chain complex activities, and mitochondrial respiration are evident on the CryABR120G hearts before the onset of detectable pathologies and development of cardiac contractile dysfunction.
Keywords: R120G‐αB‐crystallin; desmin‐related myopathy; mitochondrial dynamics; mitochondrial respiration; oxidative phosphorylation.
Conflict of interest statement
None.
Figures







References
-
- Knoll R, Hoshijima M, Hoffman HM, Person V, Lorenzen‐Schmidt I, Bang ML, Hayashi T, Shiga N, Yasukawa H, Schaper W, et al. The cardiac mechanical stretch sensor machinery involves a Z disc complex that is defective in a subset of human dilated cardiomyopathy. Cell. 2002;111:943–955. - PubMed
-
- Bennardini F, Wrzosek A, Chiesi M. Alpha B‐crystallin in cardiac tissue. Association with actin and desmin filaments. Circ Res. 1992;71:288–294. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

