Spatial heterogeneity of bacterial colonization across different gut segments following inter-species microbiota transplantation
- PMID: 33208178
- PMCID: PMC7677849
- DOI: 10.1186/s40168-020-00917-7
Spatial heterogeneity of bacterial colonization across different gut segments following inter-species microbiota transplantation
Abstract
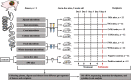
Background: The microbiota presents a compartmentalized distribution across different gut segments. Hence, the exogenous microbiota from a particular gut segment might only invade its homologous gut location during microbiota transplantation. Feces as the excreted residue contain most of the large-intestinal microbes but lack small-intestinal microbes. We speculated that whole-intestinal microbiota transplantation (WIMT), comprising jejunal, ileal, cecal, and colonic microbiota, would be more effective for reshaping the entire intestinal microbiota than conventional fecal microbiota transplantation fecal microbiota transplantation (FMT).
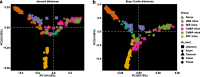
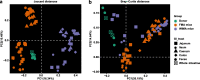
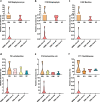
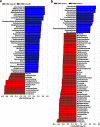
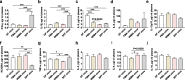
Results: We modeled the compartmentalized colonization of the gut microbiota via transplanting the microbiota from jejunum, ileum, cecum, and colon, respectively, into the germ-free mice. Transplanting jejunal or ileal microbiota induced more exogenous microbes' colonization in the small intestine (SI) of germ-free mice rather than the large intestine (LI), primarily containing Proteobacteria, Lactobacillaceae, and Cyanobacteria. Conversely, more saccharolytic anaerobes from exogenous cecal or colonic microbiota, such as Bacteroidetes, Prevotellaceae, Lachnospiraceae, and Ruminococcaceae, established in the LI of germ-free mice that received corresponding intestinal segmented microbiota transplantation. Consistent compartmentalized colonization patterns of microbial functions in the intestine of germ-free mice were also observed. Genes related to nucleotide metabolism, genetic information processing, and replication and repair were primarily enriched in small-intestinal communities, whereas genes associated with the metabolism of essential nutrients such as carbohydrates, amino acids, cofactors, and vitamins were mainly enriched in large-intestinal communities of germ-free mice. Subsequently, we compared the difference in reshaping the community structure of germ-free mice between FMT and WIMT. FMT mainly transferred LI-derived microorganisms and gene functions into the recipient intestine with sparse SI-derived microbes successfully transplanted. However, WIMT introduced more SI-derived microbes and associated microbial functions to the recipient intestine than FMT. Besides, WIMT also improved intestinal morphological development as well as reduced systematic inflammation responses of recipients compared with FMT.
Conclusions: Segmented exogenous microbiota transplantation proved the spatial heterogeneity of bacterial colonization along the gastrointestinal tract, i.e., the microbiota from one specific location selectively colonizes its homologous gut region. Given the lack of exogenous small-intestinal microbes during FMT, WIMT may be a promising alternative for conventional FMT to reconstitute the microbiota across the entire intestinal tract. Video Abstract.
Keywords: Different gut segments; Fecal microbiota transplantation; Gut microbiota; Spatial heterogeneity; Whole-intestinal microbiota transplantation.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
-
- Suez J, Zmora N, Zilberman-Schapira G, Mor U, Dori-Bachash M, Bashiardes S, Zur M, Regev-Lehavi D, Ben-Zeev Brik R, Federici S, et al. Post-antibiotic gut mucosal microbiome reconstitution is impaired by probiotics and improved by autologous FMT. Cell. 2018;174(6):1406-23.e16. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

