Capsid Structure of Leishmania RNA Virus 1
- PMID: 33208443
- PMCID: PMC7925086
- DOI: 10.1128/JVI.01957-20
Capsid Structure of Leishmania RNA Virus 1
Abstract
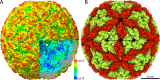
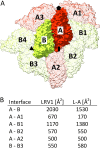
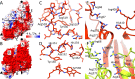
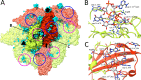
Leishmania parasites cause a variety of symptoms, including mucocutaneous leishmaniasis, which results in the destruction of the mucous membranes of the nose, mouth, and throat. The species of Leishmania carrying Leishmania RNA virus 1 (LRV1), from the family Totiviridae, are more likely to cause severe disease and are less sensitive to treatment than those that do not contain the virus. Although the importance of LRV1 for the severity of leishmaniasis was discovered a long time ago, the structure of the virus remained unknown. Here, we present a cryo-electron microscopy reconstruction of the virus-like particle of LRV1 determined to a resolution of 3.65 Å. The capsid has icosahedral symmetry and is formed by 120 copies of a capsid protein assembled in asymmetric dimers. RNA genomes of viruses from the family Totiviridae are synthetized, but not capped at the 5' end, by virus RNA polymerases. To protect viral RNAs from degradation, capsid proteins of the L-A totivirus cleave the 5' caps of host mRNAs, creating decoys to overload the cellular RNA quality control system. Capsid proteins of LRV1 form positively charged clefts, which may be the cleavage sites for the 5' cap of Leishmania mRNAs. The putative RNA binding site of LRV1 is distinct from that of the related L-A virus. The structure of the LRV1 capsid enables the rational design of compounds targeting the putative decapping site. Such inhibitors may be developed into a treatment for mucocutaneous leishmaniasis caused by LRV1-positive species of LeishmaniaIMPORTANCE Twelve million people worldwide suffer from leishmaniasis, resulting in more than 30 thousand deaths annually. The disease has several variants that differ in their symptoms. The mucocutaneous form, which leads to disintegration of the nasal septum, lips, and palate, is caused predominantly by Leishmania parasites carrying Leishmania RNA virus 1 (LRV1). Here, we present the structure of the LRV1 capsid determined using cryo-electron microscopy. Capsid proteins of a related totivirus, L-A virus, protect viral RNAs from degradation by cleaving the 5' caps of host mRNAs. Capsid proteins of LRV1 may have the same function. We show that the LRV1 capsid contains positively charged clefts that may be sites for the cleavage of mRNAs of Leishmania cells. The structure of the LRV1 capsid enables the rational design of compounds targeting the putative mRNA cleavage site. Such inhibitors may be used as treatments for mucocutaneous leishmaniasis.
Keywords: CAP-4; LRV1; Leishmania; RNA; Totiviridae; Viannia; capsid; cryo-electron microscopy; decapping; genome; leishmaniasis; mRNA; parasite; structure; uncoating; virion; virus.
Copyright © 2021 Procházková et al.
Figures



References
-
- World Health Organization. 2020. Leishmaniasis. https://www.who.int/en/news-room/fact-sheets/detail/leishmaniasis. Accessed 24 November 2020.
-
- Bruschi F, Gradoni L (ed). 2018. The leishmaniases: old neglected tropical diseases. Springer Nature, Cham, Switzerland. doi:10.1007/978-3-319-72386-0. - DOI
-
- de Oliveira Ramos Pereira L, Maretti-Mira AC, Rodrigues KM, Lima RB, de Oliveira-Neto MP, Cupolillo E, Pirmez C, Pereira de Oliveira M. 2013. Severity of tegumentary leishmaniasis is not exclusively associated with Leishmania RNA virus 1 infection in Brazil. Mem Inst Oswaldo Cruz 108:665–667. doi:10.1590/0074-0276108052013021. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

