Assessment of In Vitro and In Silico Protocols for Sequence-Based Characterization of the Human Vaginal Microbiome
- PMID: 33208514
- PMCID: PMC7677004
- DOI: 10.1128/mSphere.00448-20
Assessment of In Vitro and In Silico Protocols for Sequence-Based Characterization of the Human Vaginal Microbiome
Erratum in
-
Erratum for Hugerth et al., "Assessment of In Vitro and In Silico Protocols for Sequence-Based Characterization of the Human Vaginal Microbiome".mSphere. 2020 Dec 23;5(6):e01253-20. doi: 10.1128/mSphere.01253-20. mSphere. 2020. PMID: 33361132 Free PMC article. No abstract available.
Abstract
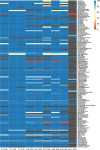
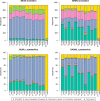
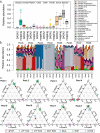
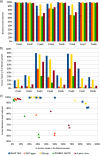
The vaginal microbiome has been connected to a wide range of health outcomes. This has led to a thriving research environment but also to the use of conflicting methodologies to study its microbial composition. Here, we systematically assessed best practices for the sequencing-based characterization of the human vaginal microbiome. As far as 16S rRNA gene sequencing is concerned, the V1-V3 region performed best in silico, but limitations of current sequencing technologies meant that the V3-V4 region performed equally well. Both approaches presented very good agreement with qPCR quantification of key taxa, provided that an appropriate bioinformatic pipeline was used. Shotgun metagenomic sequencing presents an interesting alternative to 16S rRNA gene amplification and sequencing but requires deeper sequencing and more bioinformatic expertise and infrastructure. We assessed different tools for the removal of host reads and the taxonomic annotation of metagenomic reads, including a new, easy-to-build and -use reference database of vaginal taxa. This curated database performed as well as the best-performing previously published strategies. Despite the many advantages of shotgun sequencing, none of the shotgun approaches assessed here agreed with the qPCR data as well as the 16S rRNA gene sequencing.IMPORTANCE The vaginal microbiome has been connected to various aspects of host health, including susceptibility to sexually transmitted infections as well as gynecological cancers and pregnancy outcomes. This has led to a thriving research environment but also to conflicting available methodologies, including many studies that do not report their molecular biological and bioinformatic methods in sufficient detail to be considered reproducible. This can lead to conflicting messages and delay progress from descriptive to intervention studies. By systematically assessing best practices for the characterization of the human vaginal microbiome, this study will enable past studies to be assessed more critically and assist future studies in the selection of appropriate methods for their specific research questions.
Keywords: 16S rRNA; PCR; amplicon; human microbiome; metagenomics; molecular methods; quantitative methods; vaginal microbiome.
Copyright © 2020 Hugerth et al.
Figures


Similar articles
-
Species-level classification of the vaginal microbiome.BMC Genomics. 2012;13 Suppl 8(Suppl 8):S17. doi: 10.1186/1471-2164-13-S8-S17. Epub 2012 Dec 17. BMC Genomics. 2012. PMID: 23282177 Free PMC article.
-
Metagenome-validated combined amplicon sequencing and text mining-based annotations for simultaneous profiling of bacteria and fungi: vaginal microbiota and mycobiota in healthy women.Microbiome. 2024 Dec 28;12(1):273. doi: 10.1186/s40168-024-01993-9. Microbiome. 2024. PMID: 39731160 Free PMC article.
-
Quantitative Assessment of Shotgun Metagenomics and 16S rDNA Amplicon Sequencing in the Study of Human Gut Microbiome.OMICS. 2018 Apr;22(4):248-254. doi: 10.1089/omi.2018.0013. OMICS. 2018. PMID: 29652573
-
Understanding and interpreting community sequencing measurements of the vaginal microbiome.BJOG. 2020 Jan;127(2):139-146. doi: 10.1111/1471-0528.15978. Epub 2019 Nov 1. BJOG. 2020. PMID: 31597208 Free PMC article. Review.
-
Current challenges and best-practice protocols for microbiome analysis.Brief Bioinform. 2021 Jan 18;22(1):178-193. doi: 10.1093/bib/bbz155. Brief Bioinform. 2021. PMID: 31848574 Free PMC article. Review.
Cited by
-
Organic vs. conventional: impact of cultivation treatments on the soil microbiota in the vineyard.Front Microbiol. 2023 Oct 12;14:1242267. doi: 10.3389/fmicb.2023.1242267. eCollection 2023. Front Microbiol. 2023. PMID: 37901804 Free PMC article.
-
Breast and endometrial safety of micronised progesterone versus norethisterone acetate in menopausal hormone therapy (PROBES): study protocol of a double-blind randomised controlled trial.BMJ Open. 2024 Oct 23;14(10):e082749. doi: 10.1136/bmjopen-2023-082749. BMJ Open. 2024. PMID: 39448218 Free PMC article.
-
Change in microbiota profile after vaginal estriol cream in postmenopausal women with stress incontinence.Front Microbiol. 2024 Mar 5;15:1302819. doi: 10.3389/fmicb.2024.1302819. eCollection 2024. Front Microbiol. 2024. PMID: 38505551 Free PMC article.
-
Compositional and functional differences of the vaginal microbiota of women with and without cervical dysplasia.Sci Rep. 2024 May 16;14(1):11183. doi: 10.1038/s41598-024-61942-2. Sci Rep. 2024. PMID: 38755259 Free PMC article.
-
Cohort profile: the Swedish Maternal Microbiome project (SweMaMi) - assessing the dynamic associations between the microbiome and maternal and neonatal adverse events.BMJ Open. 2022 Oct 26;12(10):e065825. doi: 10.1136/bmjopen-2022-065825. BMJ Open. 2022. PMID: 36288838 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
