Mutational Analysis of Known ALS Genes in an Italian Population-Based Cohort
- PMID: 33208543
- PMCID: PMC7905787
- DOI: 10.1212/WNL.0000000000011209
Mutational Analysis of Known ALS Genes in an Italian Population-Based Cohort
Abstract
Objective: To assess the burden of rare genetic variants and to estimate the contribution of known amyotrophic lateral sclerosis (ALS) genes in an Italian population-based cohort, we performed whole genome sequencing in 959 patients with ALS and 677 matched healthy controls.
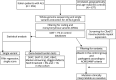
Methods: We performed genome sequencing in a population-based cohort (Piemonte and Valle d'Aosta Registry for ALS [PARALS]). A panel of 40 ALS genes was analyzed to identify potential disease-causing genetic variants and to evaluate the gene-wide burden of rare variants among our population.
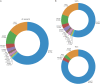
Results: A total of 959 patients with ALS were compared with 677 healthy controls from the same geographical area. Gene-wide association tests demonstrated a strong association with SOD1, whose rare variants are the second most common cause of disease after C9orf72 expansion. A lower signal was observed for TARDBP, proving that its effect on our cohort is driven by a few known causal variants. We detected rare variants in other known ALS genes that did not surpass statistical significance in gene-wise tests, thus highlighting that their contribution to disease risk in our cohort is limited.
Conclusions: We identified potential disease-causing variants in 11.9% of our patients. We identified the genes most frequently involved in our cohort and confirmed the contribution of rare variants in disease risk. Our results provide further insight into the pathologic mechanism of the disease and demonstrate the importance of genome-wide sequencing as a diagnostic tool.
© 2020 American Academy of Neurology.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
