Neoadjuvant pazopanib and molecular analysis of tissue response in renal cell carcinoma
- PMID: 33208553
- PMCID: PMC7710285
- DOI: 10.1172/jci.insight.132852
Neoadjuvant pazopanib and molecular analysis of tissue response in renal cell carcinoma
Abstract
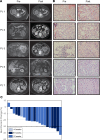
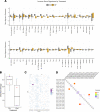
BACKGROUNDSurgery remains the frontline therapy for patients with localized clear cell renal cell carcinoma (ccRCC); however, 20%-40% recur. Angiogenesis inhibitors have improved survival in metastatic patients and may result in responses in the neoadjuvant setting. The impact of these agents on the tumor genetic heterogeneity or the immune milieu is largely unknown. This phase II study was designed to evaluate safety, response, and effect on tumor tissue of neoadjuvant pazopanib.METHODSccRCC patients with localized disease received pazopanib (800 mg daily; median 8 weeks), followed by nephrectomy. Five tumors were examined for mutations by whole exome sequencing from samples collected before therapy and at nephrectomy. These samples underwent RNA sequencing; 17 samples were available for posttreatment assessment.RESULTSTwenty-one patients were enrolled. The overall response rate was 8 of 21 (38%). No patients with progressive disease. At 1-year, response-free survival and overall survival was 83% and 89%, respectively. The most frequent grade 3 toxicity was hypertension (33%, 7 of 21). Sequencing revealed strong concordance between pre- and posttreatment samples within individual tumors, suggesting tumors harbor stable core profiles. However, a reduction in private mutations followed treatment, suggesting a selective process favoring enrichment of driver mutations.CONCLUSIONNeoadjuvant pazopanib is safe and active in ccRCC. Future genomic analyses may enable the segregation of driver and passenger mutations. Furthermore, tumor infiltrating immune cells persist during therapy, suggesting that pazopanib can be combined with immune checkpoint inhibitors without dampening the immune response.FUNDINGSupport was provided by Novartis and GlaxoSmithKline as part of an investigator-initiated study.
Keywords: Cancer; Clinical Trials; Expression profiling; Oncology; Urology.
Conflict of interest statement
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

