Disease-associated gut microbiome and metabolome changes in patients with chronic obstructive pulmonary disease
- PMID: 33208745
- PMCID: PMC7676259
- DOI: 10.1038/s41467-020-19701-0
Disease-associated gut microbiome and metabolome changes in patients with chronic obstructive pulmonary disease
Abstract
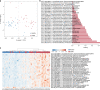
Chronic obstructive pulmonary disease (COPD) is the third commonest cause of death globally, and manifests as a progressive inflammatory lung disease with no curative treatment. The lung microbiome contributes to COPD progression, but the function of the gut microbiome remains unclear. Here we examine the faecal microbiome and metabolome of COPD patients and healthy controls, finding 146 bacterial species differing between the two groups. Several species, including Streptococcus sp000187445, Streptococcus vestibularis and multiple members of the family Lachnospiraceae, also correlate with reduced lung function. Untargeted metabolomics identifies a COPD signature comprising 46% lipid, 20% xenobiotic and 20% amino acid related metabolites. Furthermore, we describe a disease-associated network connecting Streptococcus parasanguinis_B with COPD-associated metabolites, including N-acetylglutamate and its analogue N-carbamoylglutamate. While correlative, our results suggest that the faecal microbiome and metabolome of COPD patients are distinct from those of healthy individuals, and may thus aid in the search for biomarkers for COPD.
Conflict of interest statement
P.H. is a co-founder of Microba Life Sciences Limited, and D.L.A.W. is currently an employee of Microba. The remaining authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

