Defining HPV-specific B cell responses in patients with head and neck cancer
- PMID: 33208941
- PMCID: PMC9462833
- DOI: 10.1038/s41586-020-2931-3
Defining HPV-specific B cell responses in patients with head and neck cancer
Abstract
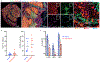
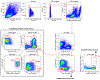
Tumours often contain B cells and plasma cells but the antigen specificity of these intratumoral B cells is not well understood1-8. Here we show that human papillomavirus (HPV)-specific B cell responses are detectable in samples from patients with HPV-positive head and neck cancers, with active production of HPV-specific IgG antibodies in situ. HPV-specific antibody secreting cells (ASCs) were present in the tumour microenvironment, with minimal bystander recruitment of influenza-specific cells, suggesting a localized and antigen-specific ASC response. HPV-specific ASC responses correlated with titres of plasma IgG and were directed against the HPV proteins E2, E6 and E7, with the most dominant response against E2. Using intratumoral B cells and plasma cells, we generated several HPV-specific human monoclonal antibodies, which exhibited a high degree of somatic hypermutation, consistent with chronic antigen exposure. Single-cell RNA sequencing analyses detected activated B cells, germinal centre B cells and ASCs within the tumour microenvironment. Compared with the tumour parenchyma, B cells and ASCs were preferentially localized in the tumour stroma, with well-formed clusters of activated B cells indicating ongoing germinal centre reactions. Overall, we show that antigen-specific activated and germinal centre B cells as well as plasma cells can be found in the tumour microenvironment. Our findings provide a better understanding of humoral immune responses in human cancer and suggest that tumour-infiltrating B cells could be harnessed for the development of therapeutic agents.
© 2020. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Figures

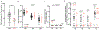





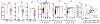


Comment in
-
A refined view of tumour-associated B cells.Nat Rev Cancer. 2021 Feb;21(2):67. doi: 10.1038/s41568-020-00328-8. Nat Rev Cancer. 2021. PMID: 33340031 No abstract available.
References
Additional references for methods section
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

