MFSD12 mediates the import of cysteine into melanosomes and lysosomes
- PMID: 33208952
- PMCID: PMC7770032
- DOI: 10.1038/s41586-020-2937-x
MFSD12 mediates the import of cysteine into melanosomes and lysosomes
Abstract
Dozens of genes contribute to the wide variation in human pigmentation. Many of these genes encode proteins that localize to the melanosome-the organelle, related to the lysosome, that synthesizes pigment-but have unclear functions1,2. Here we describe MelanoIP, a method for rapidly isolating melanosomes and profiling their labile metabolite contents. We use this method to study MFSD12, a transmembrane protein of unknown molecular function that, when suppressed, causes darker pigmentation in mice and humans3,4. We find that MFSD12 is required to maintain normal levels of cystine-the oxidized dimer of cysteine-in melanosomes, and to produce cysteinyldopas, the precursors of pheomelanin synthesis made in melanosomes via cysteine oxidation5,6. Tracing and biochemical analyses show that MFSD12 is necessary for the import of cysteine into melanosomes and, in non-pigmented cells, lysosomes. Indeed, loss of MFSD12 reduced the accumulation of cystine in lysosomes of fibroblasts from patients with cystinosis, a lysosomal-storage disease caused by inactivation of the lysosomal cystine exporter cystinosin7-9. Thus, MFSD12 is an essential component of the cysteine importer for melanosomes and lysosomes.
Conflict of interest statement
COMPETING INTEREST
The authors declare the following competing interests: C.H.A, A.K.T., and D.M.S are listed on a patent application based on the work described here.
Figures
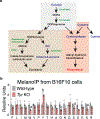






References
-
- Basrur V. et al. Proteomic Analysis of Early Melanosomes: Identification of Novel Melanosomal Proteins. J. Proteome Res. 2, 69–79 (2003). - PubMed
-
- Sturm RA Molecular genetics of human pigmentation diversity. Human Molecular Genetics 18, R9–R17 (2009). - PubMed
-
- D’Alba L. & Shawkey MD Melanosomes: Biogenesis, Properties, and Evolution of an Ancient Organelle. Physiological Reviews 99, 1–19 (2019). - PubMed
METHODS REFERENCES
-
- Guan X, Hoffman B, Dwivedi C. & Matthees DP A simultaneous liquid chromatography/mass spectrometric assay of glutathione, cysteine, homocysteine and their disulfides in biological samples. Journal of Pharmaceutical and Biomedical Analysis 31, 251–261 (2003). - PubMed
-
- Martin GB et al. Development of a mass spectrometry method for the determination of a melanoma biomarker, 5-S-cysteinyldopa, in human plasma using solid phase extraction for sample clean-up. Journal of Chromatography A 1156, 141–148 (2007). - PubMed
-
- Ito S. & Prota GMA A facile one-step synthesis of cysteinyldopas using mushroom tyrosinase. Experientia 33, 1118–1119 (1977). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

