Gene expression profiling reveals insights into infant immunological and febrile responses to group B meningococcal vaccine
- PMID: 33210468
- PMCID: PMC7674973
- DOI: 10.15252/msb.20209888
Gene expression profiling reveals insights into infant immunological and febrile responses to group B meningococcal vaccine
Abstract
Neisseria meningitidis is a major cause of meningitis and septicaemia. A MenB vaccine (4CMenB) was licensed by the European Medicines Agency in January 2013. Here we describe the blood transcriptome and proteome following infant immunisations with or without concomitant 4CMenB, to gain insight into the molecular mechanisms underlying post-vaccination reactogenicity and immunogenicity. Infants were randomised to receive control immunisations (PCV13 and DTaP-IPV-Hib) with or without 4CMenB at 2 and 4 months of age. Blood gene expression and plasma proteins were measured prior to, then 4 h, 24 h, 3 days or 7 days post-vaccination. 4CMenB vaccination was associated with increased expression of ENTPD7 and increased concentrations of 4 plasma proteins: CRP, G-CSF, IL-1RA and IL-6. Post-vaccination fever was associated with increased expression of SELL, involved in neutrophil recruitment. A murine model dissecting the vaccine components found the concomitant regimen to be associated with increased gene perturbation compared with 4CMenB vaccine alone with enhancement of pathways such as interleukin-3, -5 and GM-CSF signalling. Finally, we present transcriptomic profiles predictive of immunological and febrile responses following 4CMenB vaccine.
Trial registration: ClinicalTrials.gov NCT02080559.
Keywords: paediatrics; proteomics; systems biology; transcriptomics; vaccines.
© 2020 The Authors. Published under the terms of the CC BY 4.0 license.
Conflict of interest statement
AJP reports grants from Okairos outside the submitted work, and AJP is Chair of UK Department of Health and Social Care's Joint Committee on Vaccination and Immunisation, and the EMA scientific advisory group on vaccines and is a member of the World Health Organisation's Strategic Advisory Group of Experts on Immunisation. MDS acts as a Chief/Principal Investigator on clinical trials funded by vaccine manufacturers including Glaxosmithkline, Novavax, Medimmune, MCM, Pfizer and Janssen. These studies are conducted on behalf of the University of Oxford and MDS receives no personal financial benefit. MVP is a member of the Portuguese National Immunisation Technical Advisory Group (Comissão Técnica de Vacinação da Direcção Geral de Saúde).
Figures

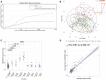
Kaplan‐Meier to first fever (≥ 38°C) episode within 24 h of receiving vaccines administered at 4 months of age; control n = 88 and 4CMenB n = 89.
Principal component (PC) analysis of RNA‐sequencing data (14,837 genes, n = 253) from all study time points. The lines connect participant's pre‐ and post‐vaccination samples. The ellipses are the two‐dimensional (PC1 and PC2) 95% confidence intervals for each study time point. A contribution plot (top right) displays the genes contributing most to PC1/PC2, i.e. 0.15 implies 0.15% contribution of that variable to the principal components displayed.
Plotted are the CIBERSORTx neutrophil fractions from whole blood RNA‐sequencing data, with median and interquartile range. P‐values were determined from a two‐sample Wilcoxon rank sum test. The number of individuals in each group is display in the x‐axis.
Spearman's rank correlation between neutrophil counts measure by full blood counts (n = 142) and those estimated by CIBERSORTx, using the LM22 signature matrix.
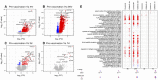
Volcano plot highlighting differentially expressed genes (DEGs, false discovery rate [FDR] < 0.01; red upregulated and blue downregulated) at each study time point versus pre‐vaccination (4 months of age) 4 h post‐vaccination (719 DEGs, n = 28). P‐values were obtained from the moderated t‐statistic, after adjustment for multiple testing (Benjamini and Hochberg's method). The top 10 genes, ranked by FDR, are labelled.
Same as (A) but 24 h post‐vaccination (5,553 DEGs, n = 31).
Same as (A) but 3 days post‐vaccination (159 DEGs, n = 30).
Same as (A) but 7 days post‐vaccination (6 DEGs, n = 36).
Modular signature induced following infant vaccination. Enriched modules (FDR < 0.001) are displayed. Segments of the pie charts represent the proportion of upregulated (red) and downregulated (blue) genes (absolute fold change > 1.25). Enrichment P‐values were derived from a hypergeometric test, after adjustment for multiple testing (Benjamini and Hochberg's method).
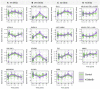
Top genes differentially regulated between vaccine groups 4 h post‐vaccination. Note, only genes differentially expressed in either study group (concomitant 4CMenB or control vaccines alone) were included in intergroup analysis. The plotted lines are the LOESS (locally estimated scatterplot smoothing) regression curves with the 95% confidence intervals in grey. The FDR was derived by comparing fold changes in gene expression in the control group with the test group, from baseline to the time point designated, and is reported if statistically significant (FDR < 0.05). P‐values were obtained from the moderated t‐statistic, after adjustment for multiple testing (Benjamini and Hochberg's method). Pre‐vaccination samples n = 125, 4 h samples n = 28, 24 h samples n = 31, 3 day sample n = 30, 7 day samples n = 36. The expression E value is the gene expression value derived from the voom‐limma workflow (Law et al, 2014).
Same as (A) but top genes differentially regulated between vaccine groups 24 h post‐vaccination.
Same as (A) but top genes differentially regulated between vaccine groups 3 days post‐vaccination.
Same as (A) but top genes differentially regulated between vaccine groups 7 days post‐vaccination.
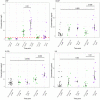

- A, B
Agreement plot. Red = differentially expressed (DE) in febrile infants, blue = DE in afebrile infants, purple = DE in febrile and afebrile infants.
- C
Exemplar genes differentially expressed at 4 and 24 h, respectively, between infants who experienced a fever within 24 h of vaccination and afebrile infants. The plotted lines are the LOESS (locally estimated scatterplot smoothing) regression curves with the 95% confidence intervals in grey. The false discovery rate (FDR) was derived by comparing fold changes in gene expression between infants who experienced post‐vaccination fever and those who remained afebrile. P‐values were obtained from the moderated t‐statistic, after adjustment for multiple testing (Benjamini and Hochberg's method). Pre‐vaccination samples n = 125, 4 h samples n = 28, 24 h samples n = 31, 3 day sample n = 30, 7 day samples n = 36.
- D
Performance of predictive model built with sparse distance weighted discrimination (sdwd) algorithm to predict fever following concomitant 4CMenB vaccine.
- E
Variable importance score of the features from the sdwd model.
- F
Expression levels (Limma E value) of the top three transcripts from the sdwd model in the individuals that develop fever (n = 33, red circles) and those that don't develop fever (n = 21, blue circles).
- G–I
Support vector regression (SVR) performance of model to predict post‐vaccination MenB‐specific SBA titres, (G) training dataset (n = 36), (H) performance of model in the test dataset (n = 9), I) the top five genes ranked by importance from the SVR model.


References
-
- Andrews NJ, Walker WT, Finn A, Heath PT, Collinson AC, Pollard AJ, Snape MD, Faust SN, Waight PA, Hoschler K et al (2011) Predictors of immune response and reactogenicity to AS03B‐adjuvanted split virion and non‐adjuvanted whole virion H1N1 (2009) pandemic influenza vaccines. Vaccine 29: 7913–7919 - PubMed
-
- Bellman RE (1957) Dynamic programming. Princeton University Press, New York, NY:
-
- Bendall LJ, Bradstock KF (2014) G‐CSF: From granulopoietic stimulant to bone marrow stem cell mobilizing agent. Cytokine Growth Factor Rev 25: 355–367 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

